JET2 Viewer
Pre-computed interfaces from JET2
1A8R - hydrolase
gtp cyclohydrolase i (h112s mutant) in complex with gtp
NOTE: Use your mouse to drag, rotate, and zoom in and out of the structure. Help.
Download: 1A8R.zip
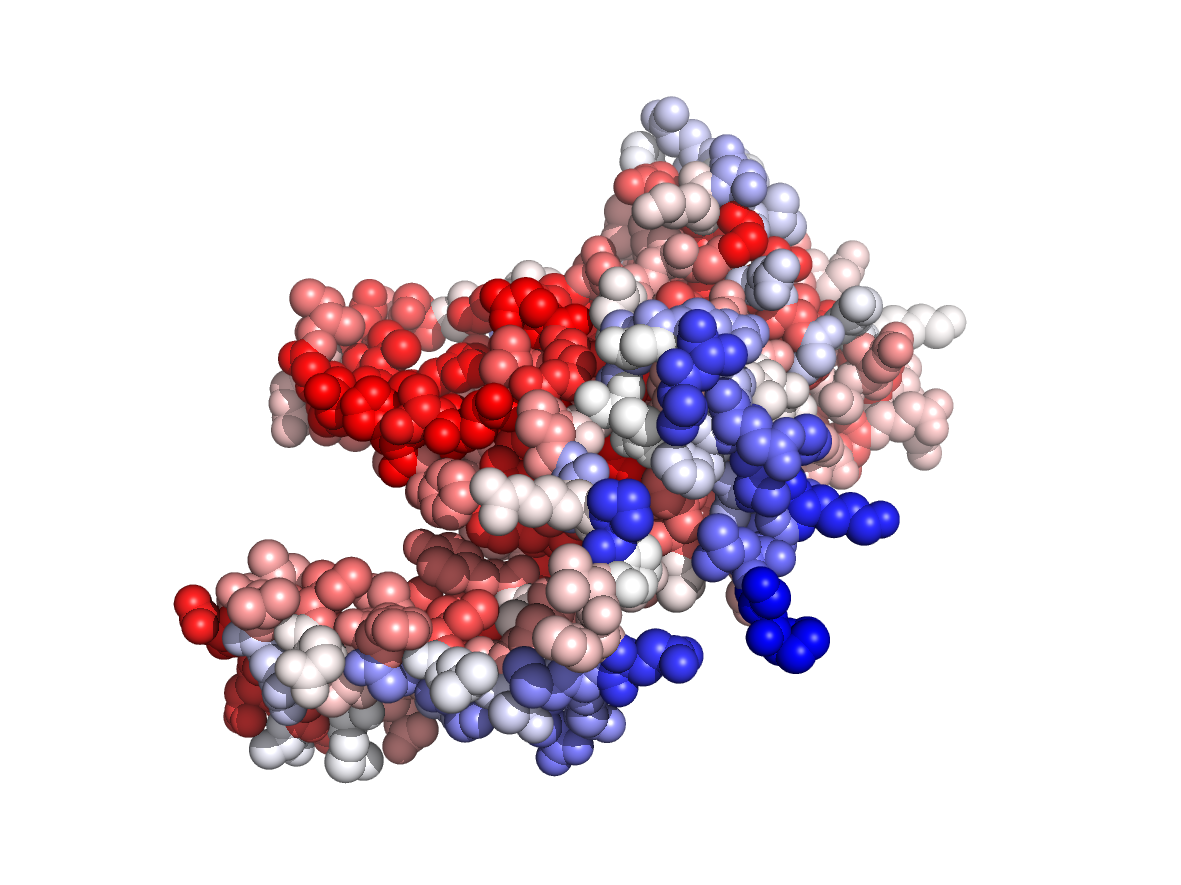
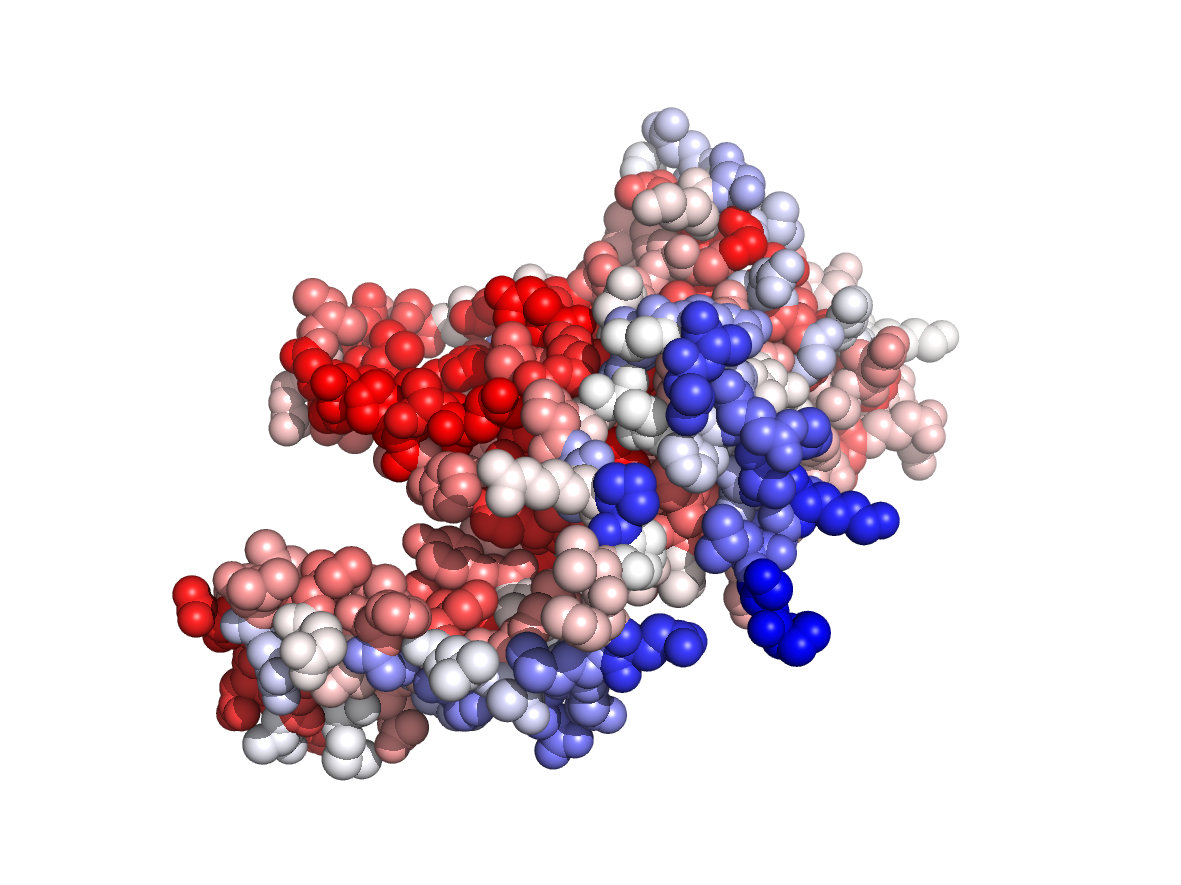
PDB Structure: 1A8R chain A
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
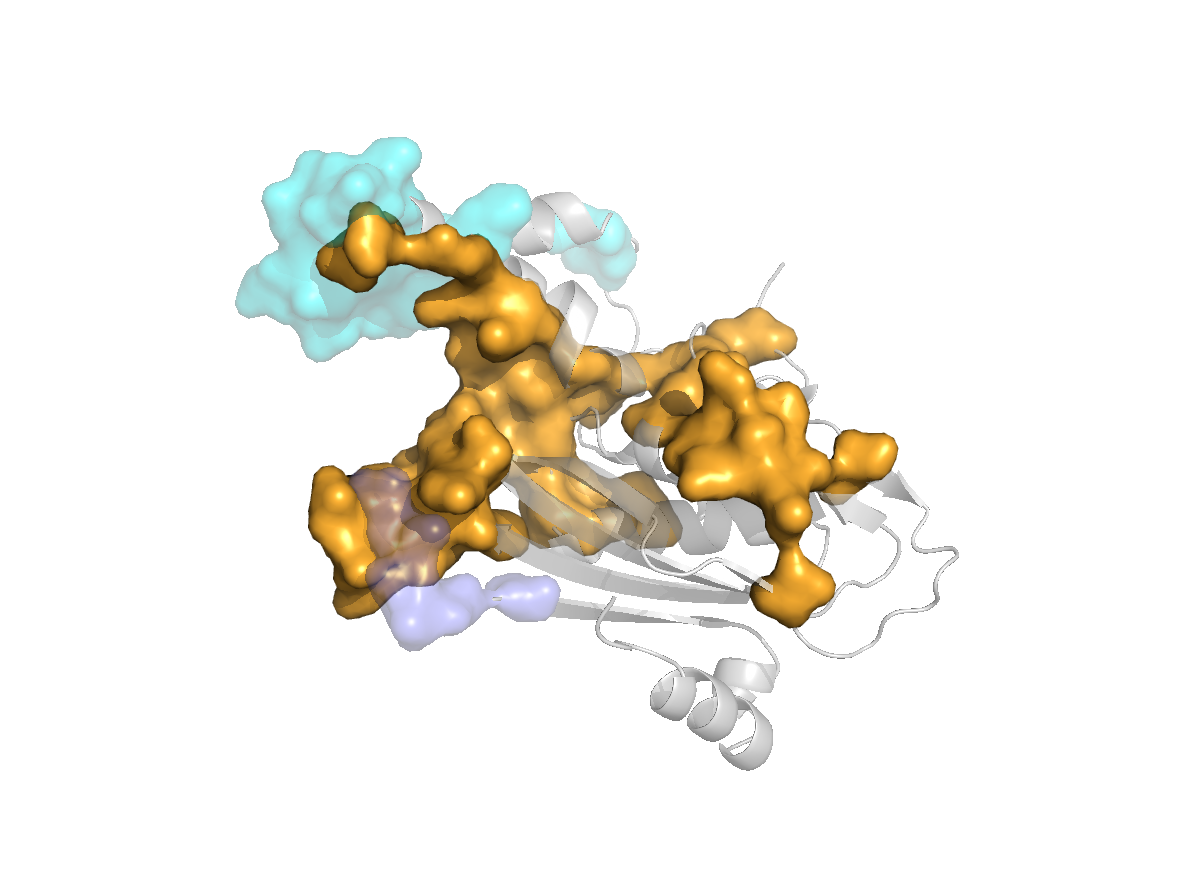 site |
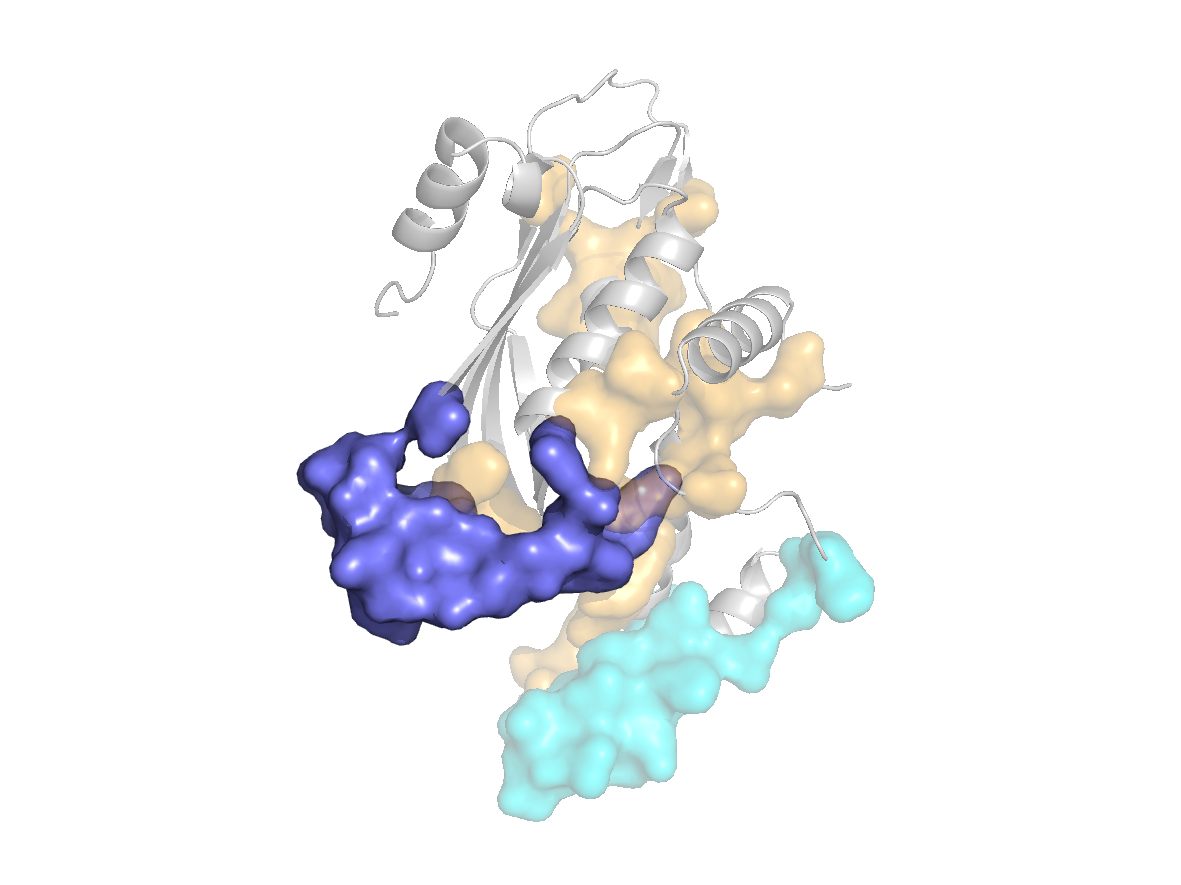 site |
 site |
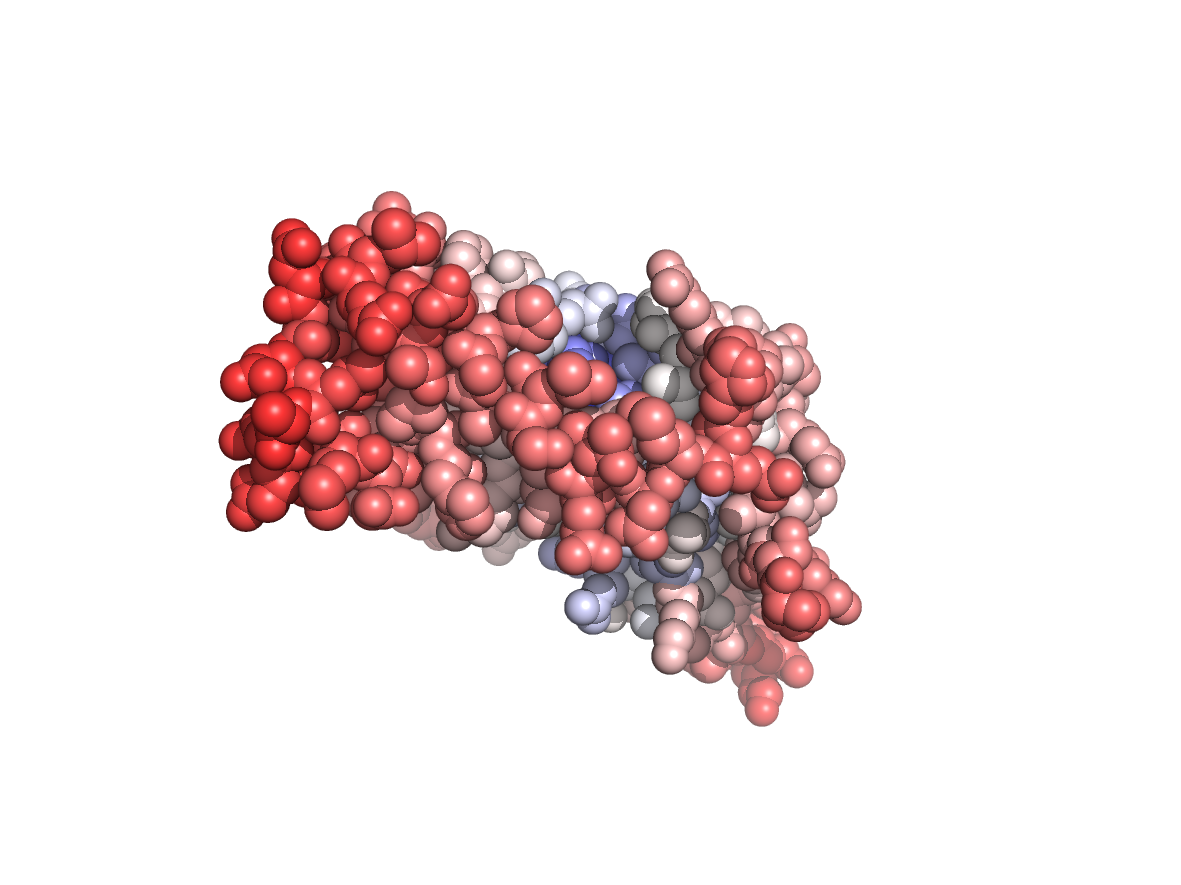
Evolutionary conservation - TJET |
Physico-chemical properties - PC |
Circular variance - CV |
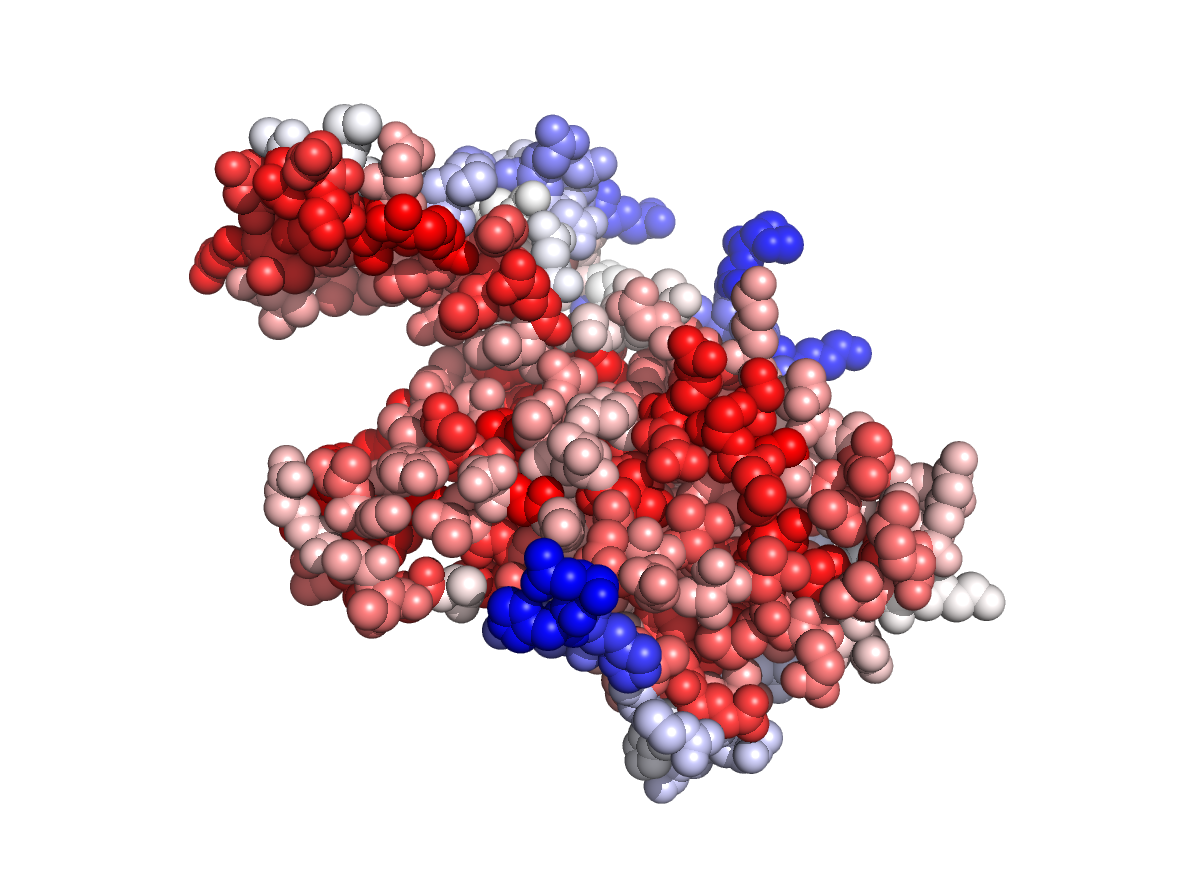 |
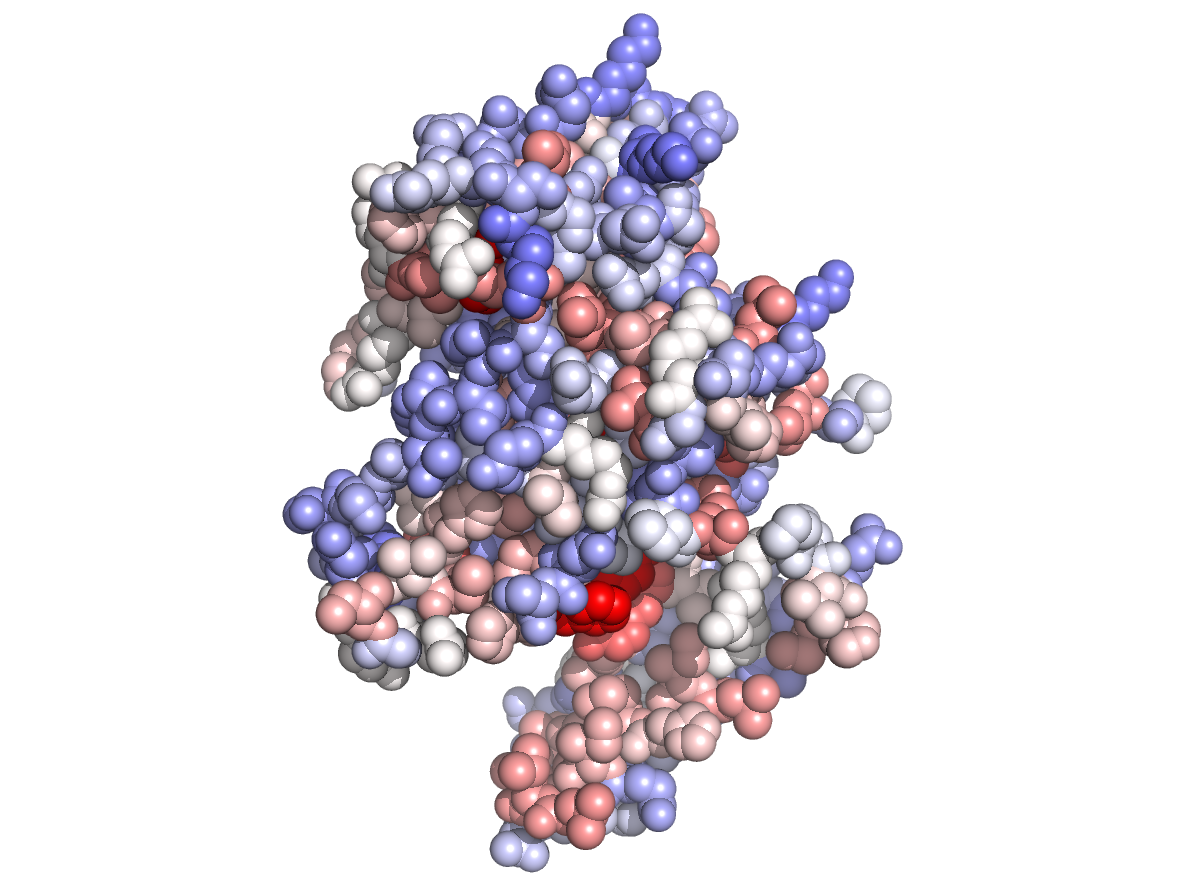 |
 |
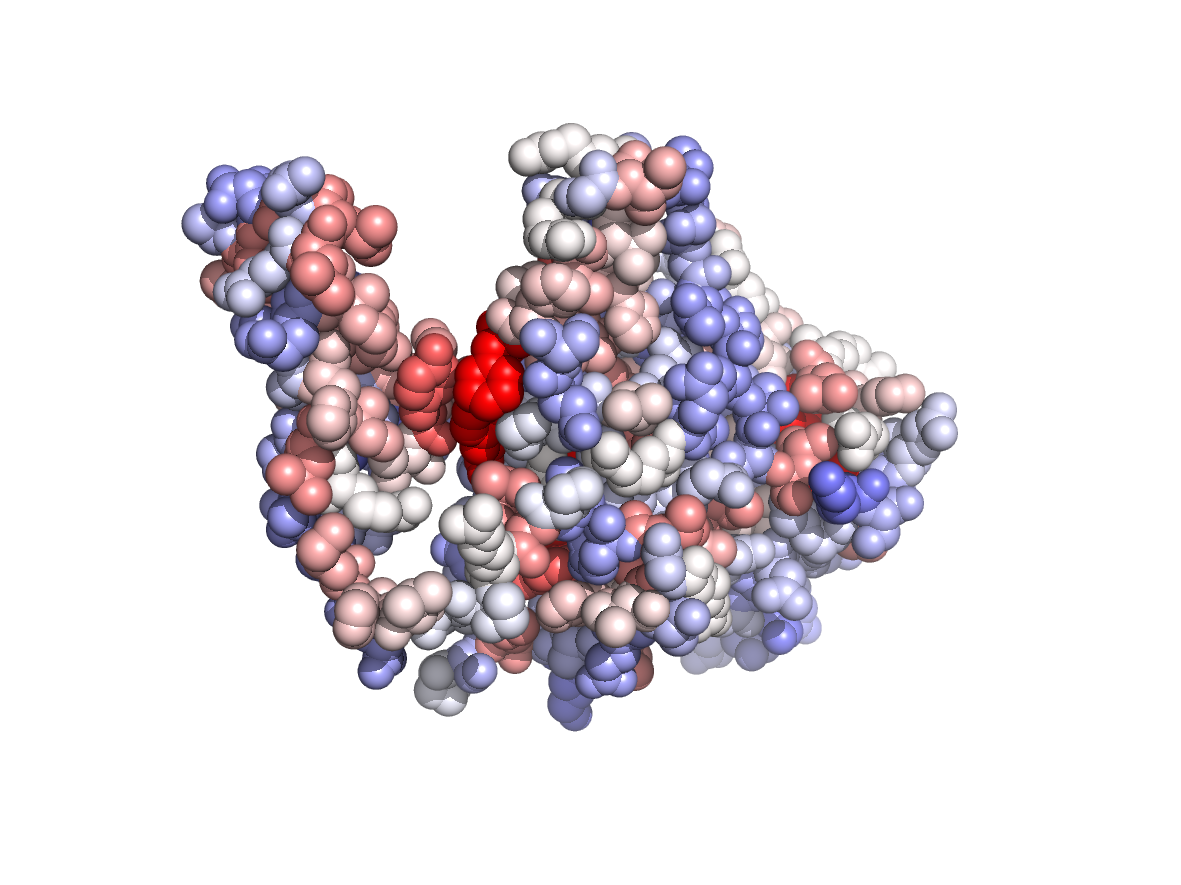
PDB Structure: 1A8R chain B
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
 site |
 site |
 site |
Evolutionary conservation - TJET |
Physico-chemical properties - PC |
Circular variance - CV |
 |
 |
 |
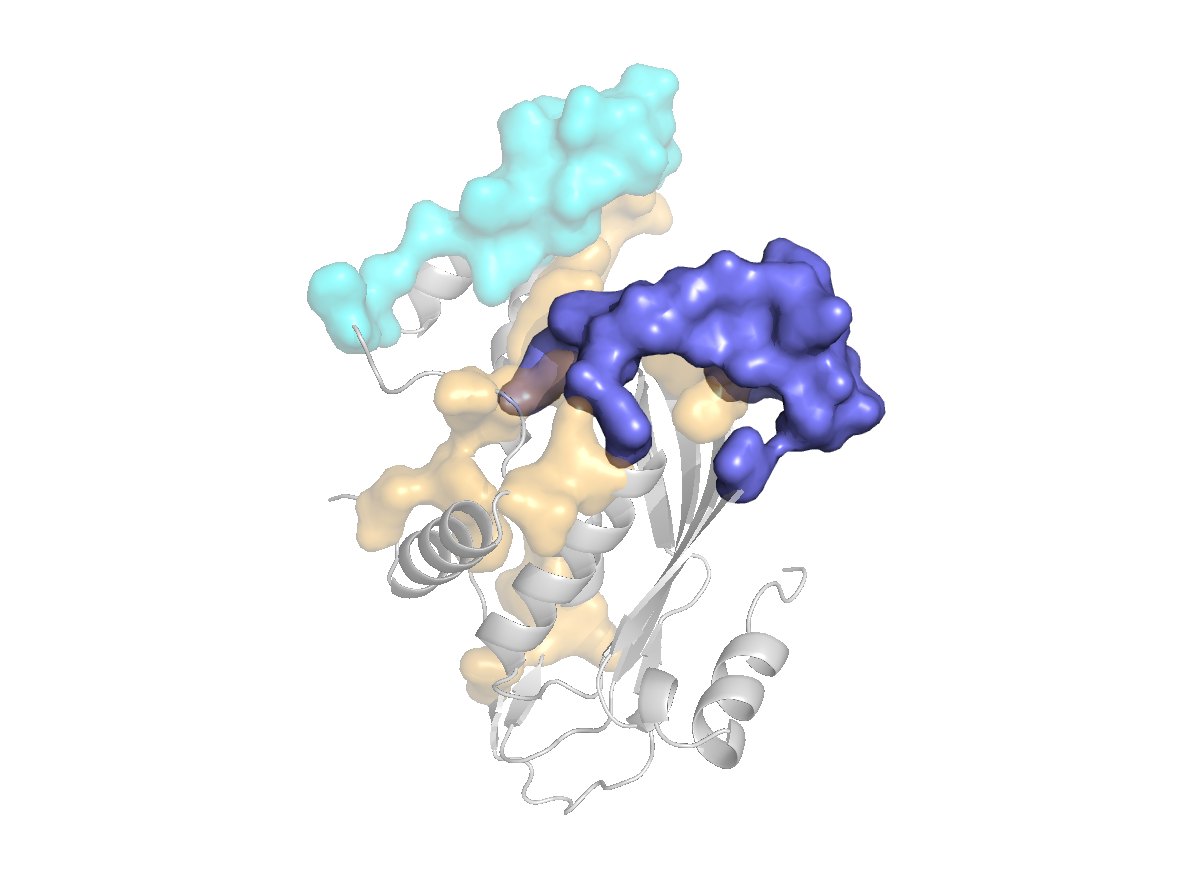
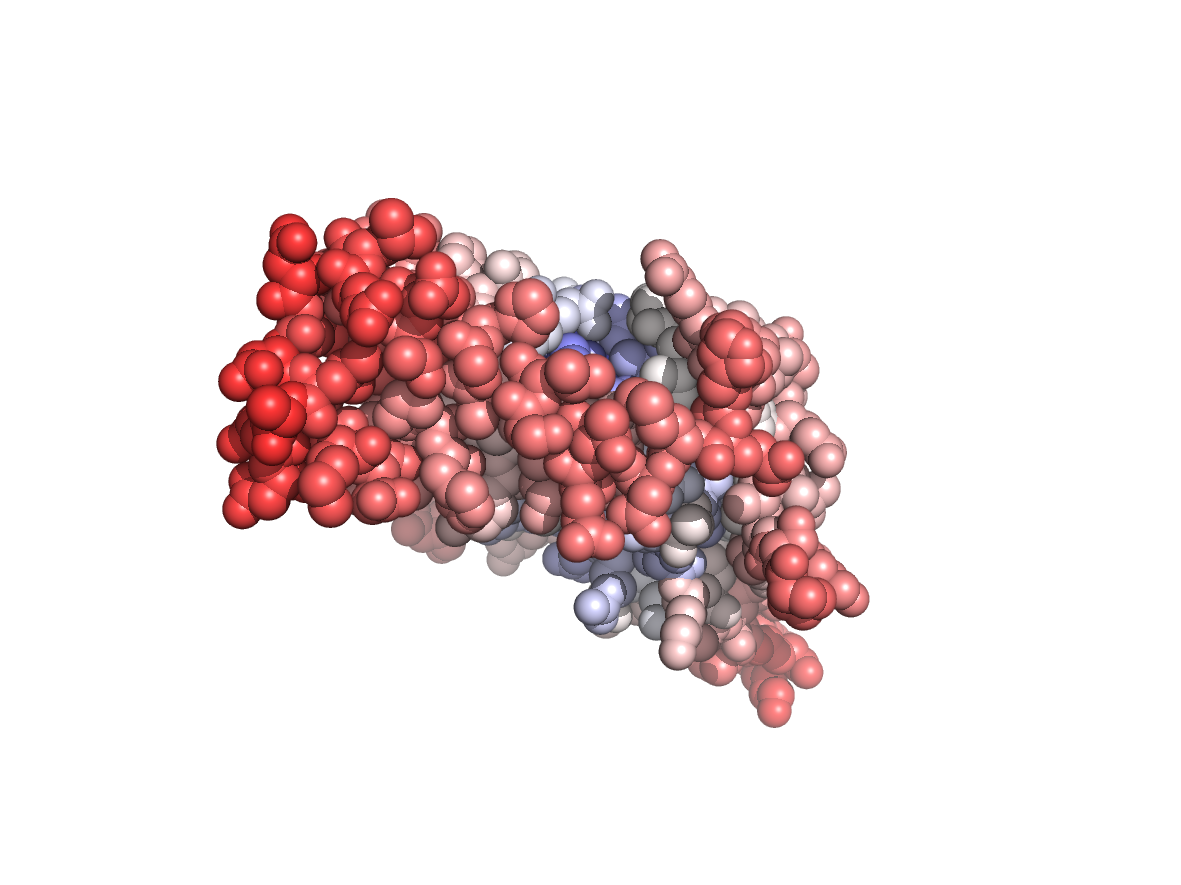
PDB Structure: 1A8R chain C
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
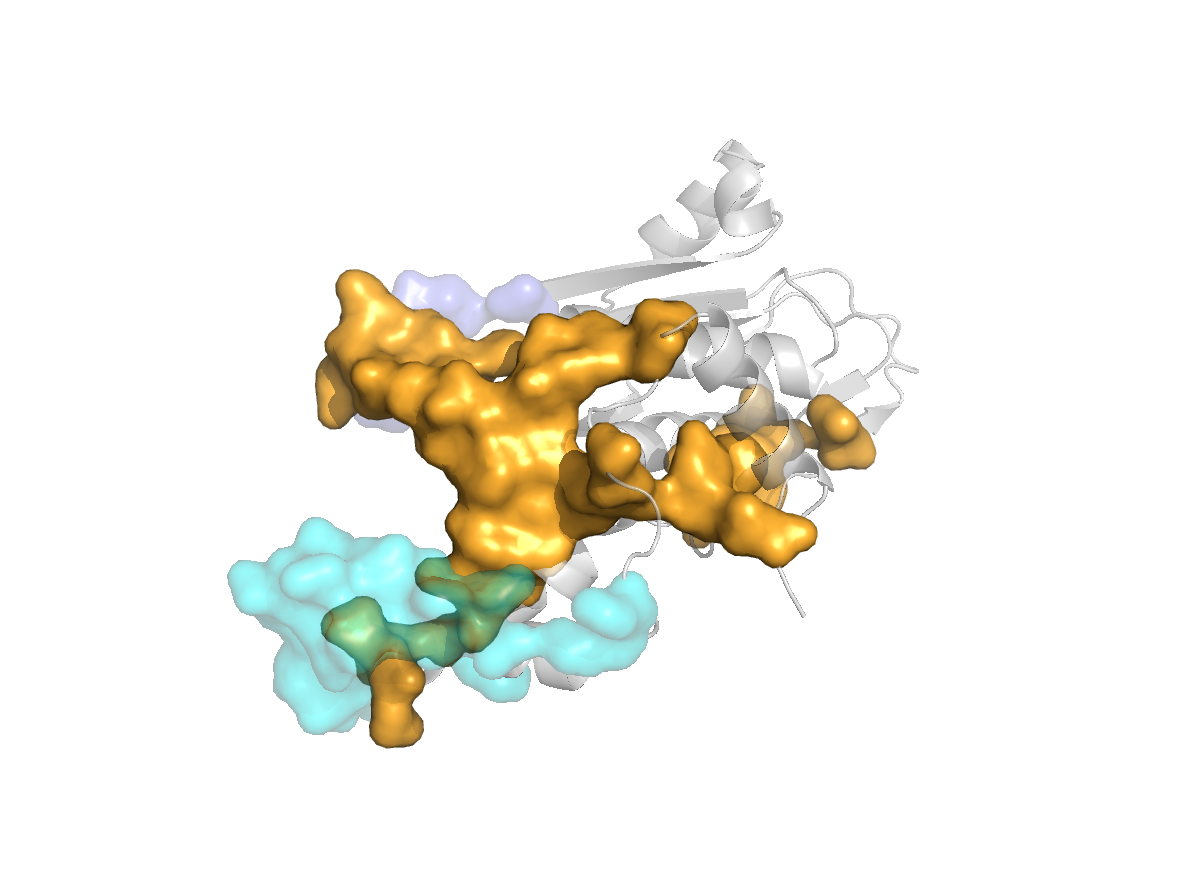 site |
 site |
 site |
Evolutionary conservation - TJET |
Physico-chemical properties - PC |
Circular variance - CV |
 |
 |
 |
PDB Structure: 1A8R chain D
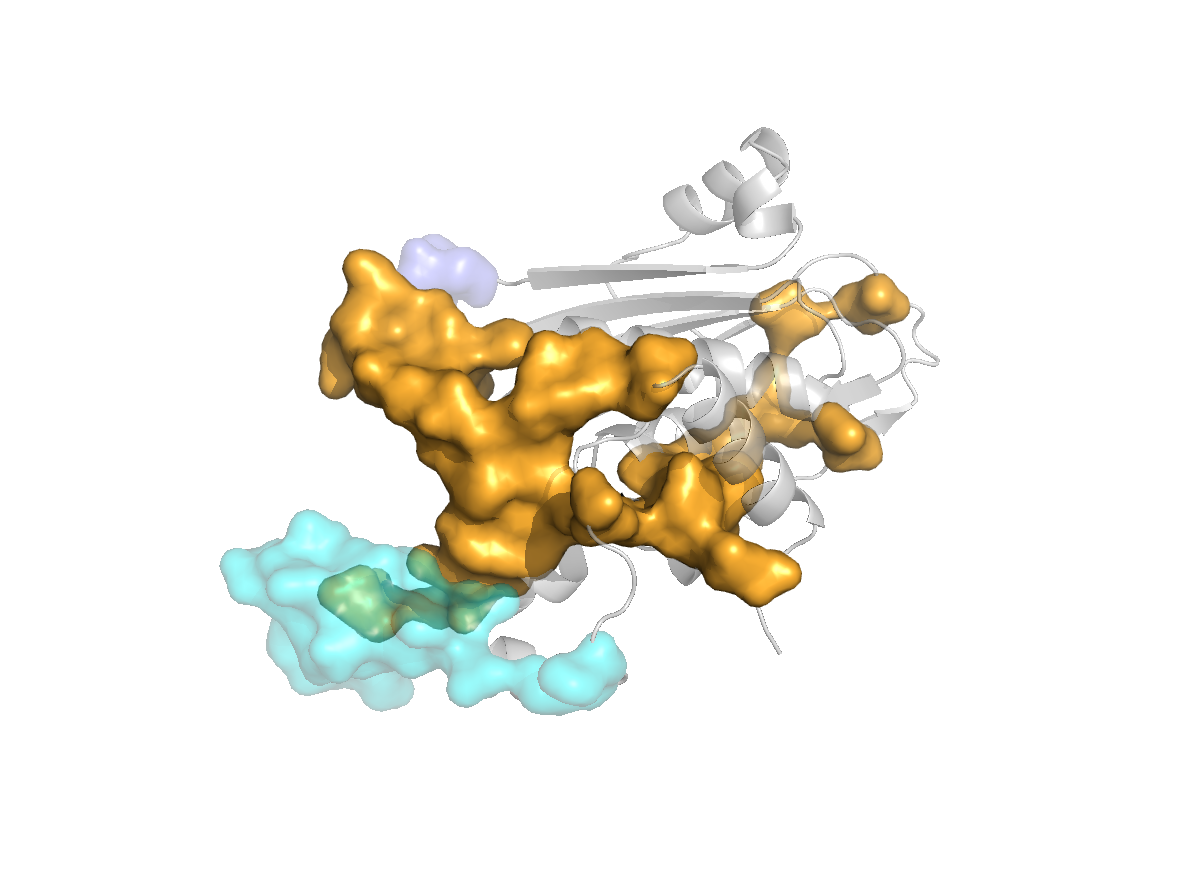
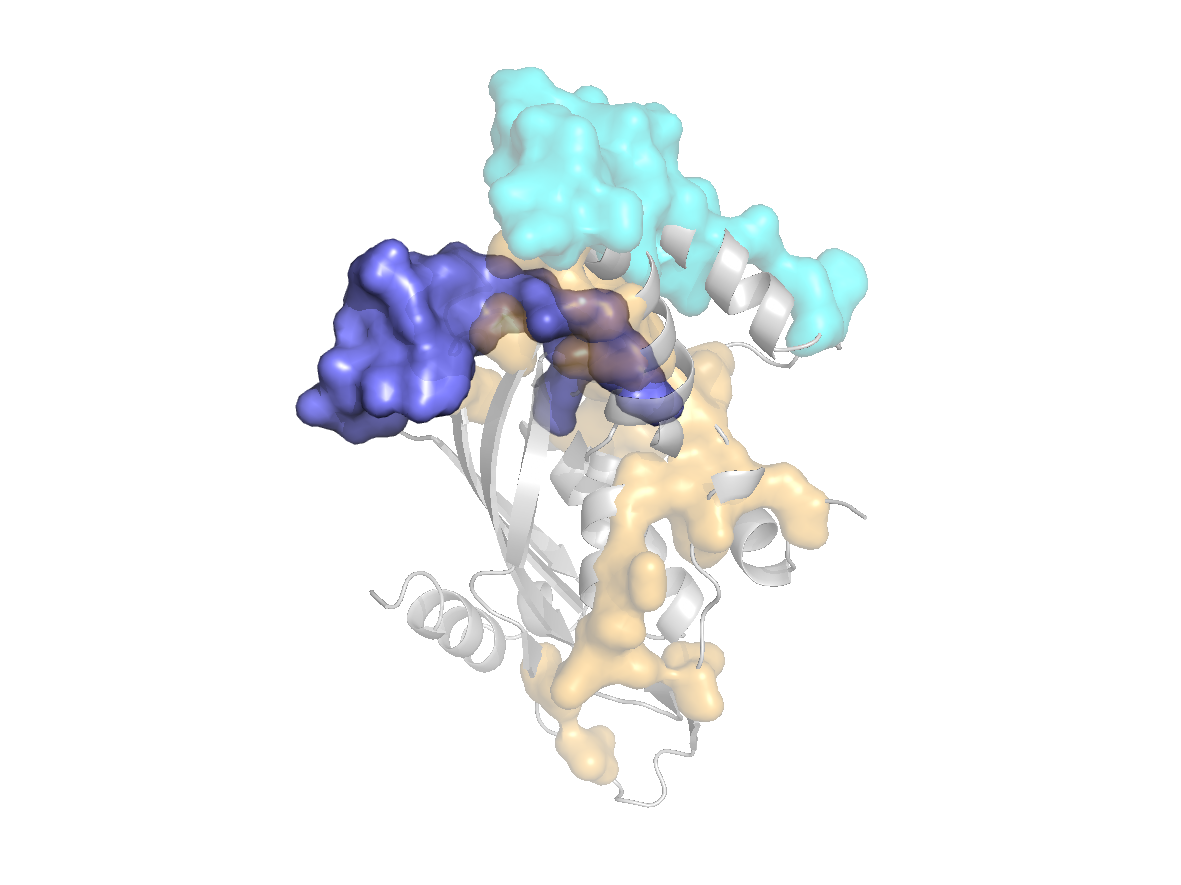
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
 site |
 site |
 site |
Evolutionary conservation - TJET |
Physico-chemical properties - PC |
Circular variance - CV |
 |
 |
 |
PDB Structure: 1A8R chain E
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
 site |
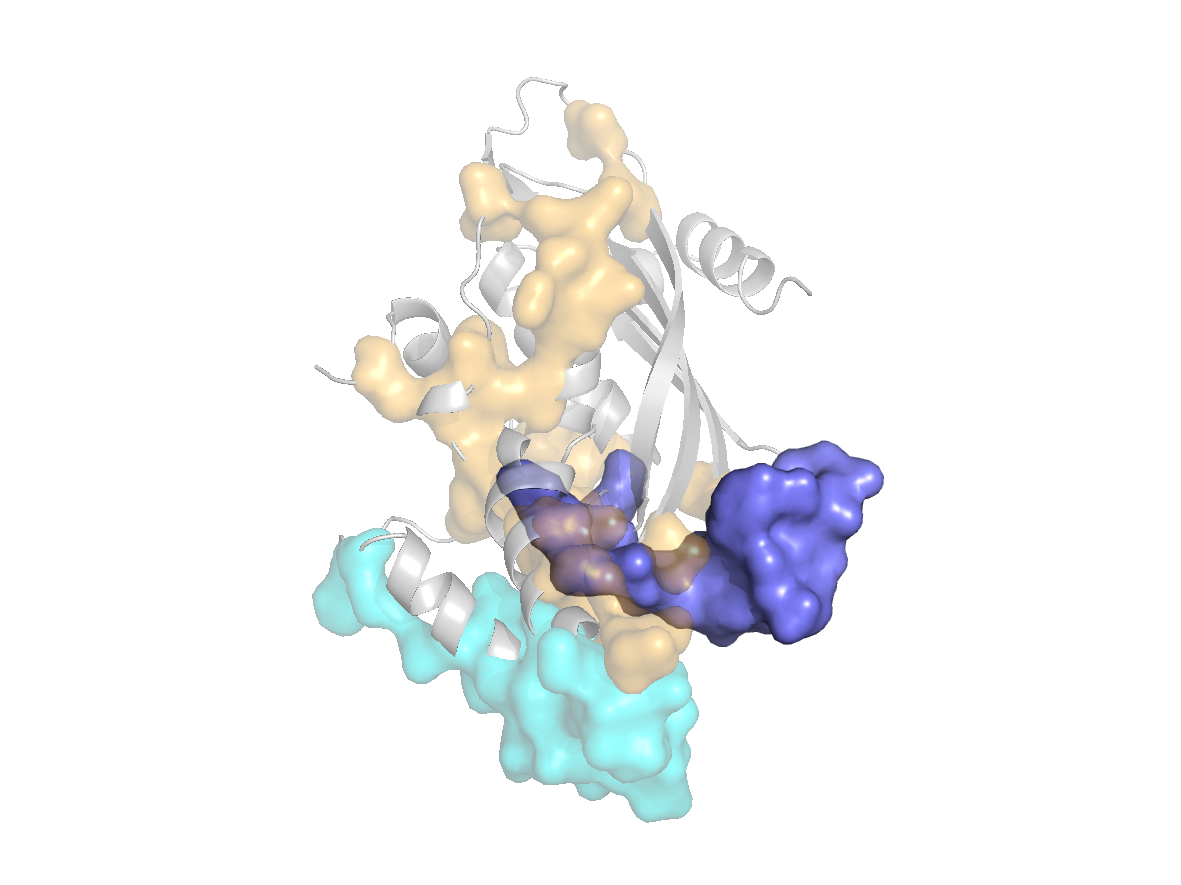 site |
 site |
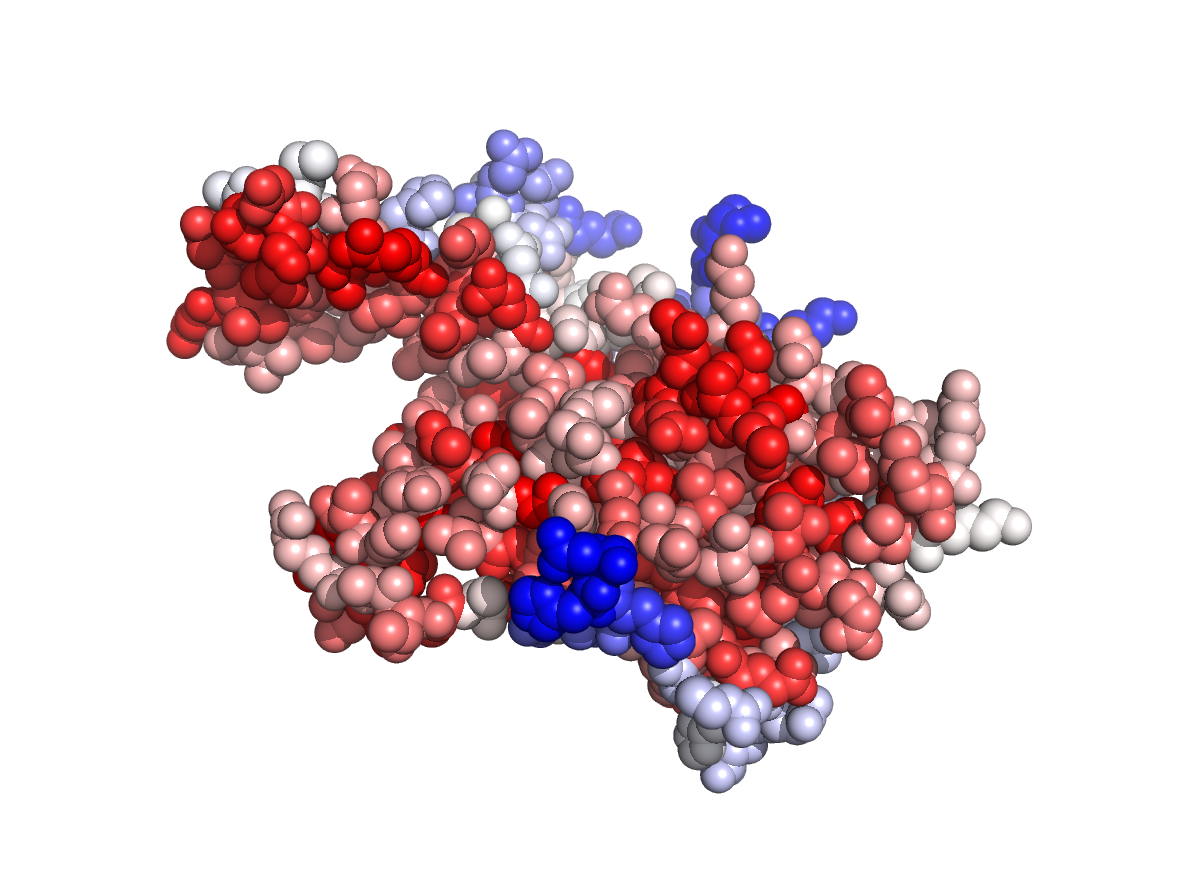
Evolutionary conservation - TJET |
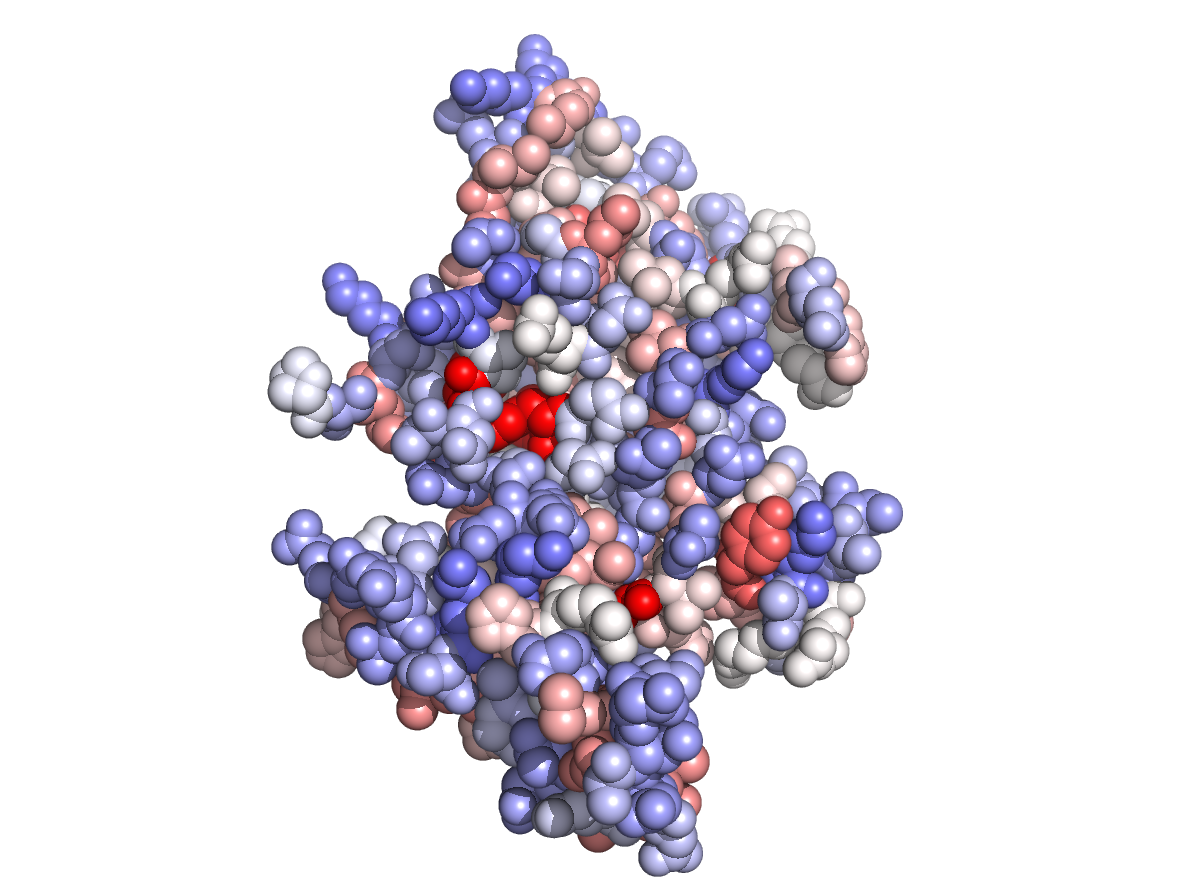
Physico-chemical properties - PC |
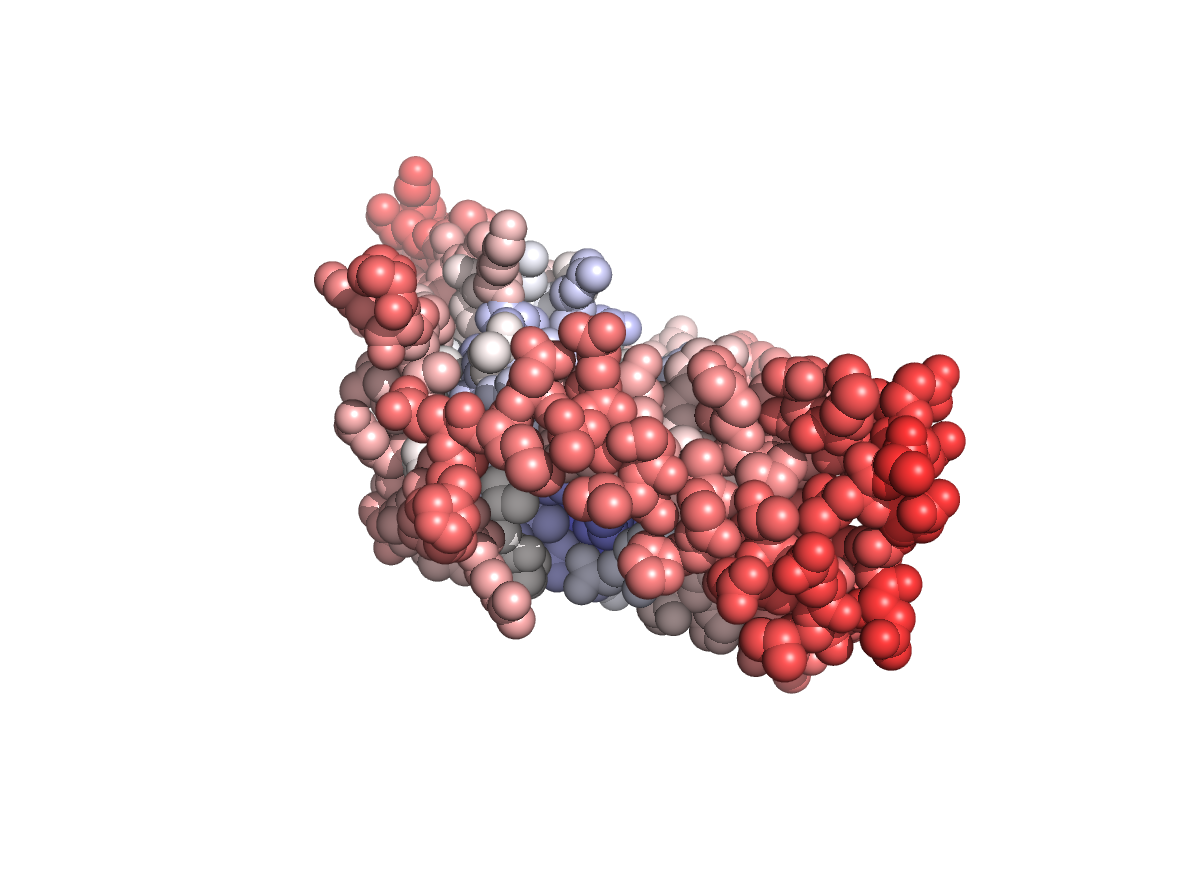
Circular variance - CV |
 |
 |
 |
PDB Structure: 1A8R chain F
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
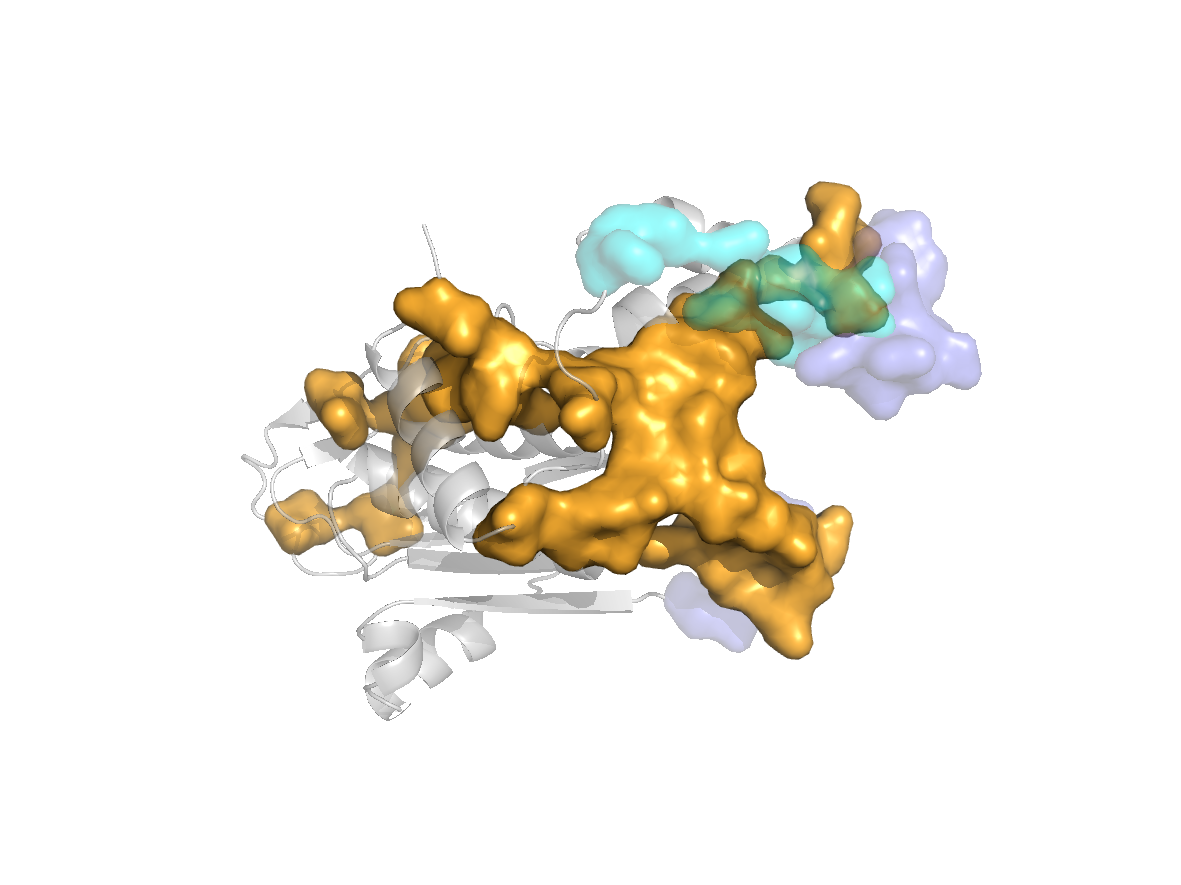 site |
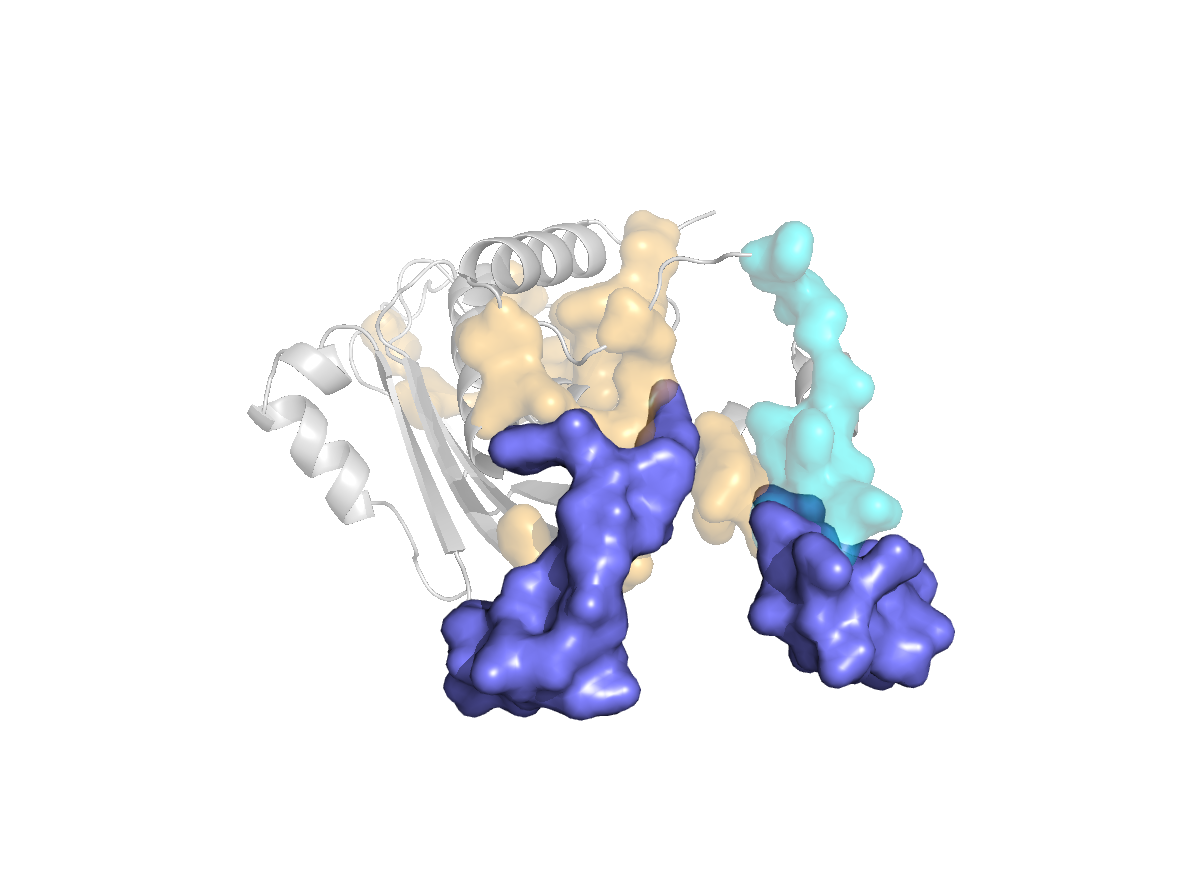 site |
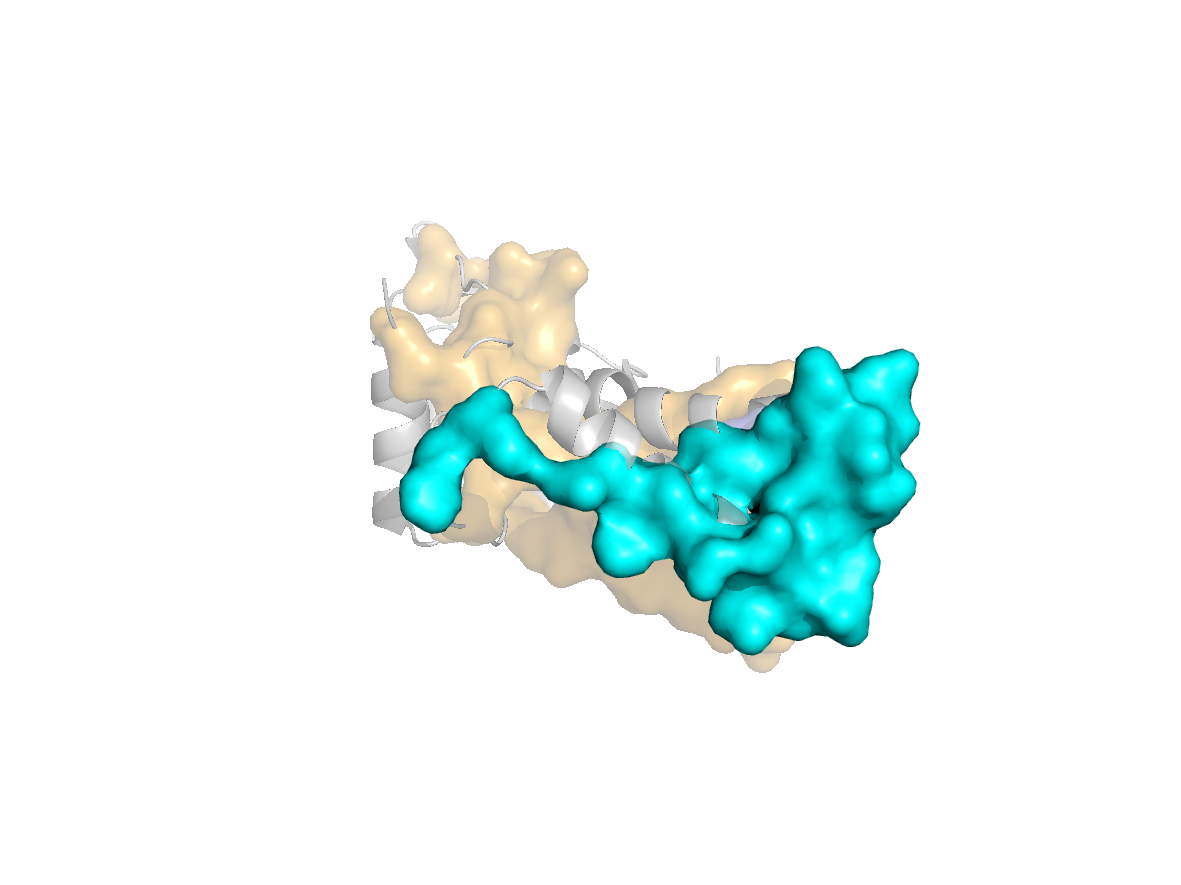 site |
Evolutionary conservation - TJET |
Physico-chemical properties - PC |
Circular variance - CV |
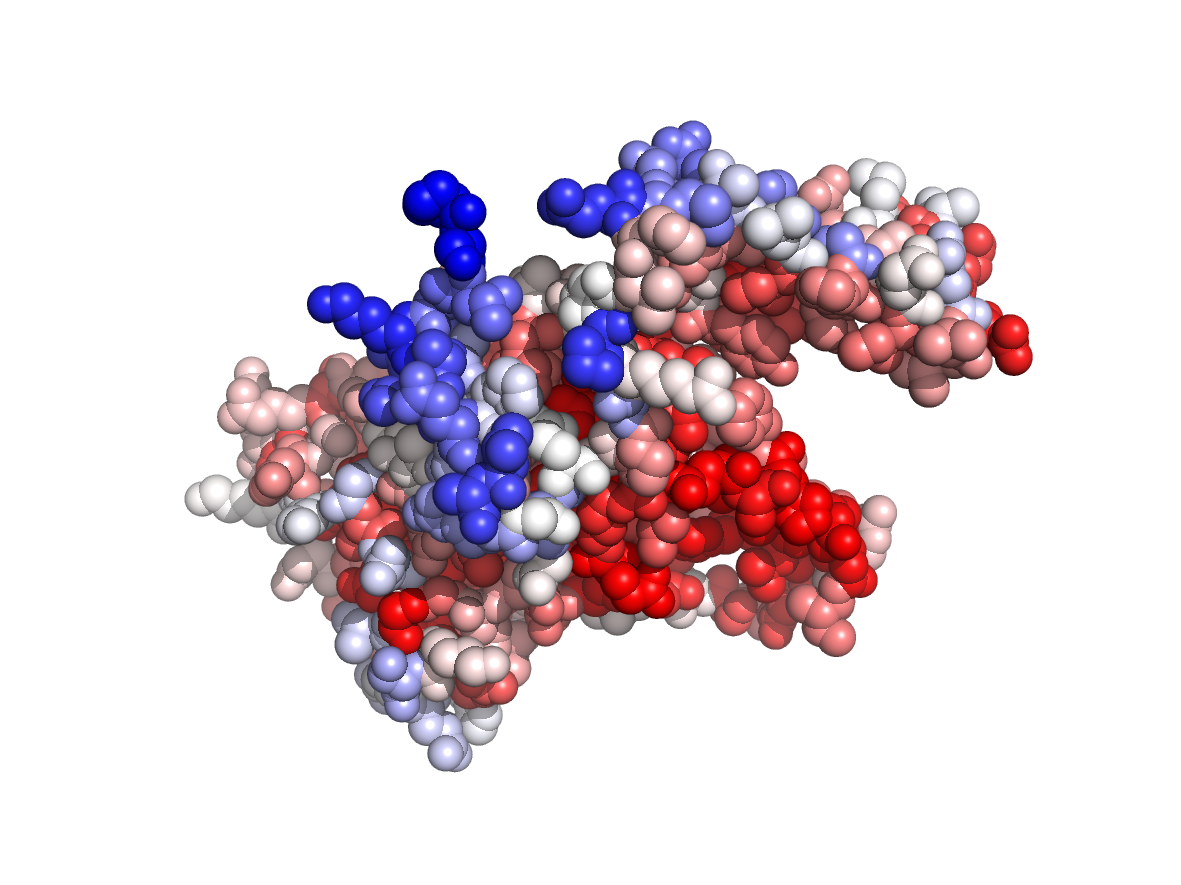 |
 |
 |
PDB Structure: 1A8R chain G
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
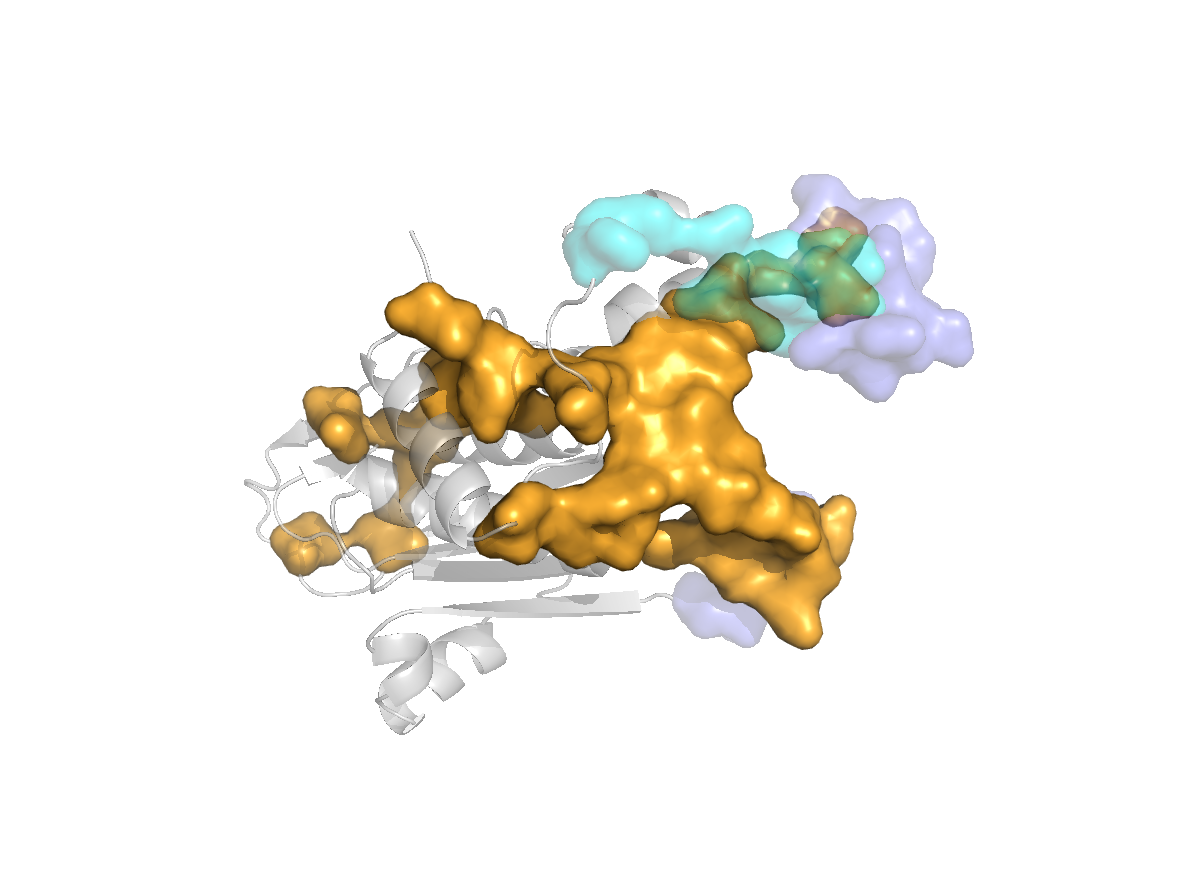 site |
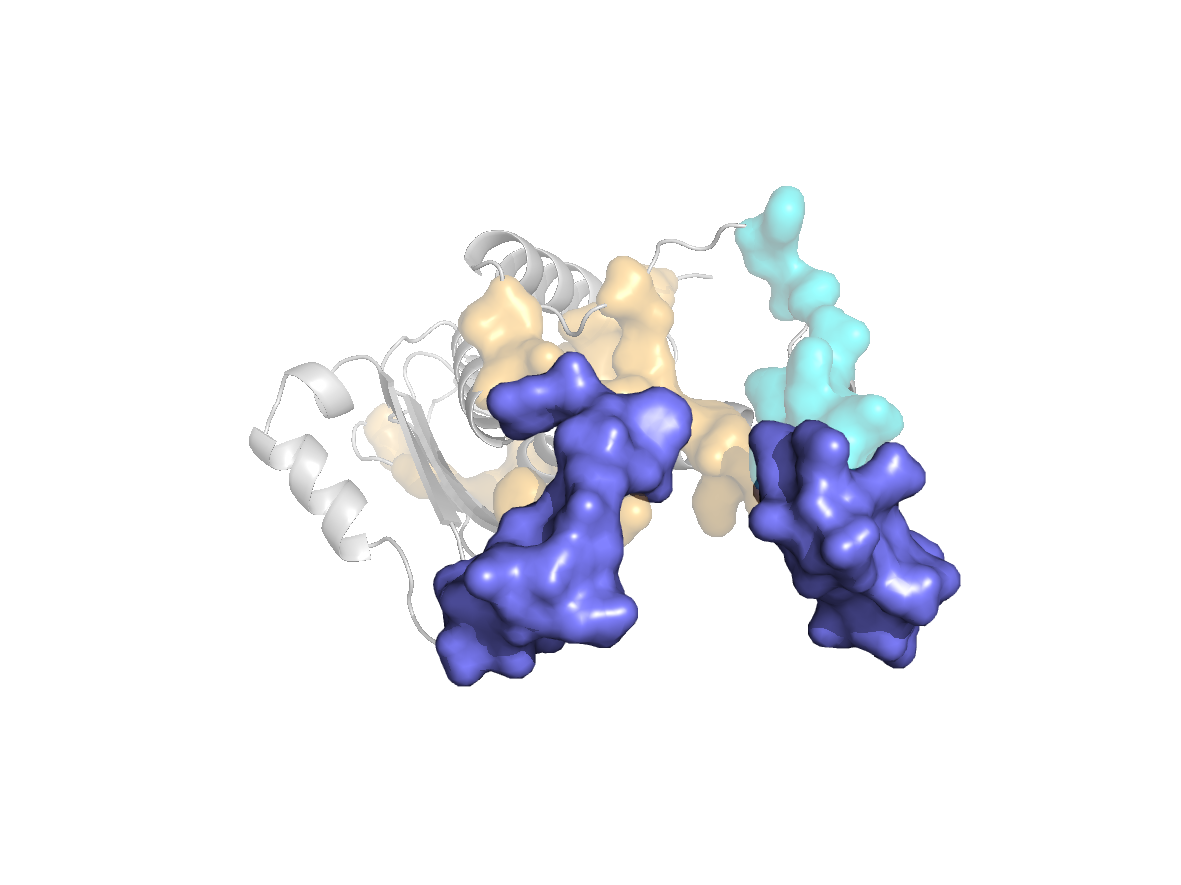 site |
 site |
Evolutionary conservation - TJET |
Physico-chemical properties - PC |
Circular variance - CV |
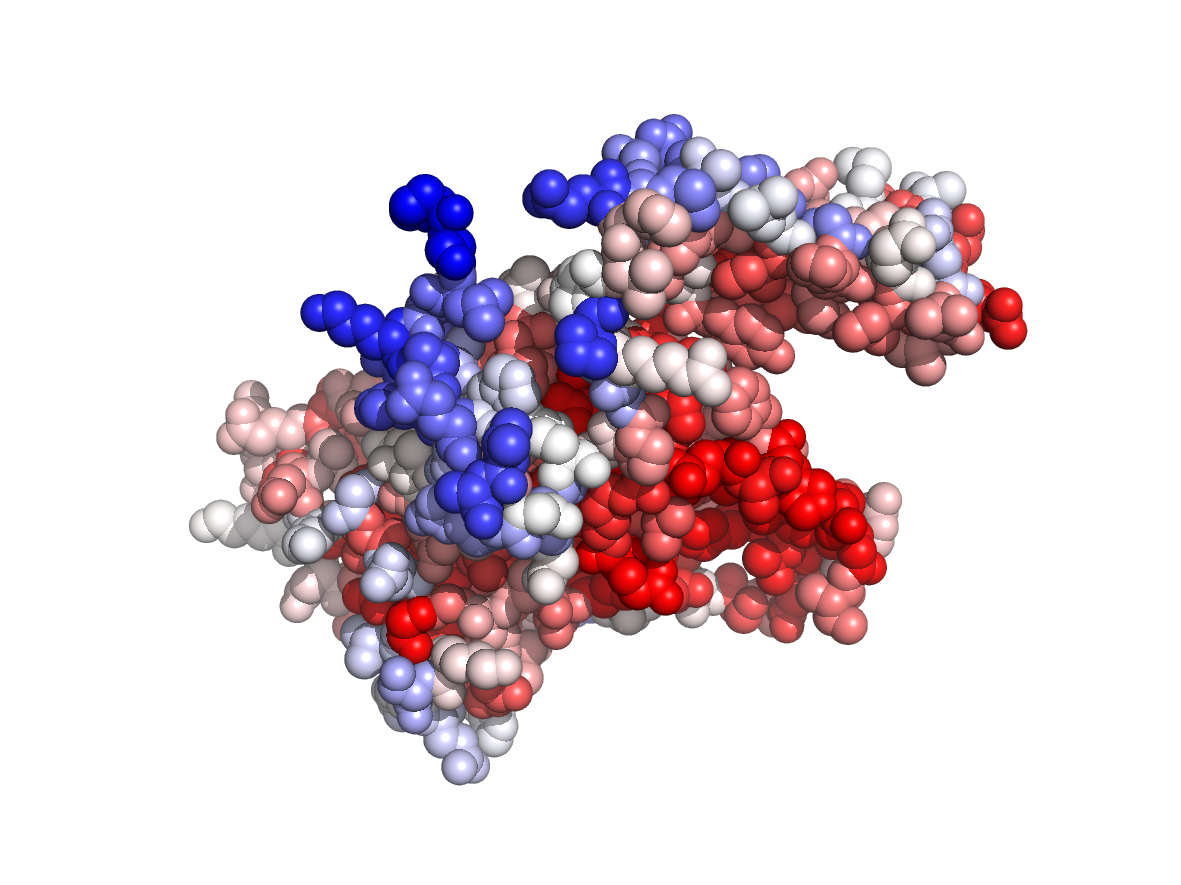 |
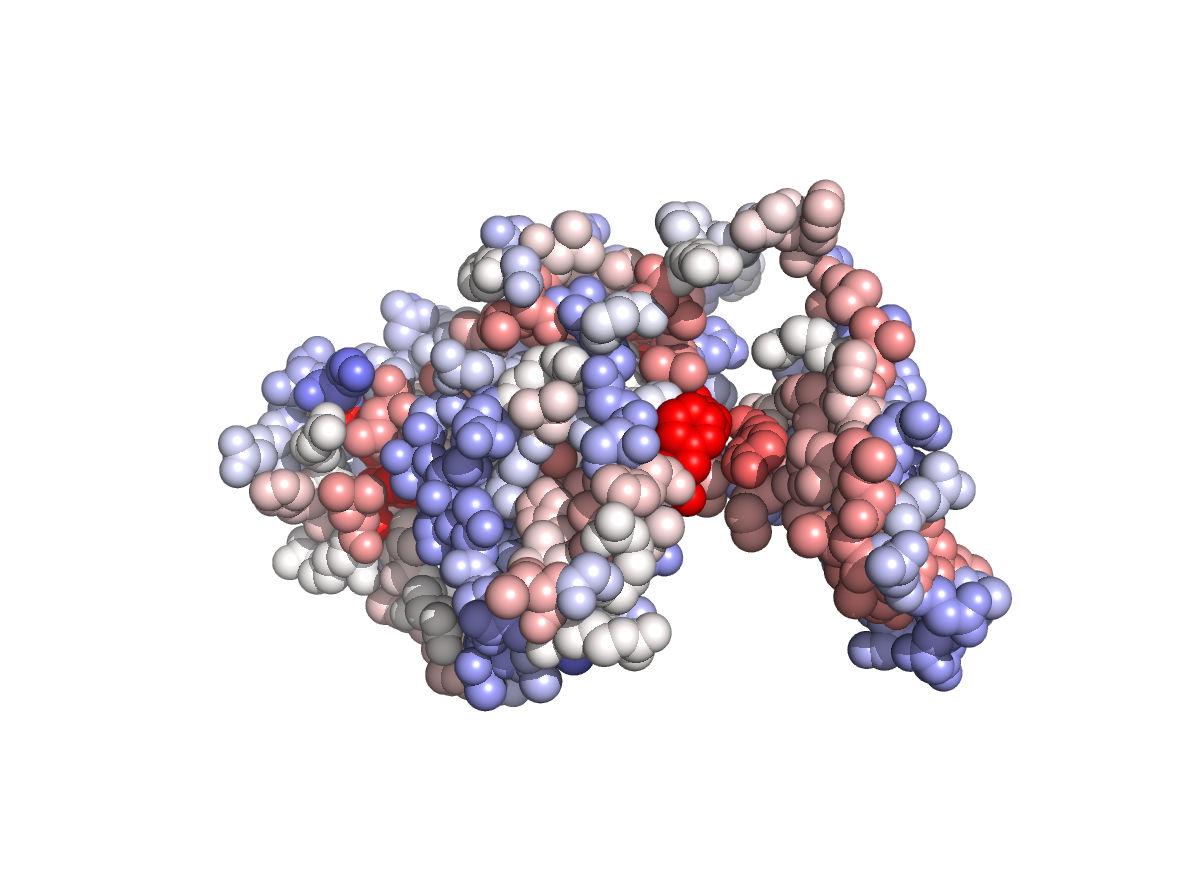 |
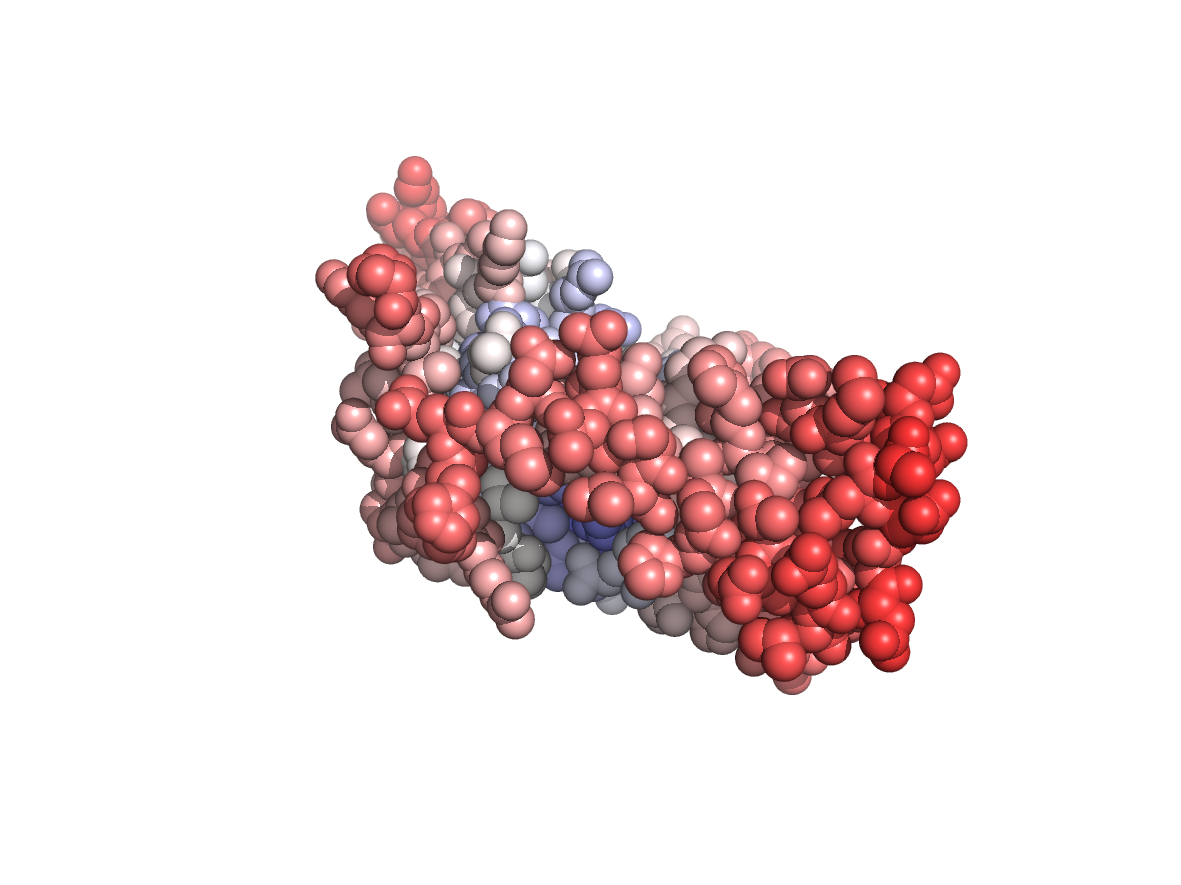 |
PDB Structure: 1A8R chain H
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
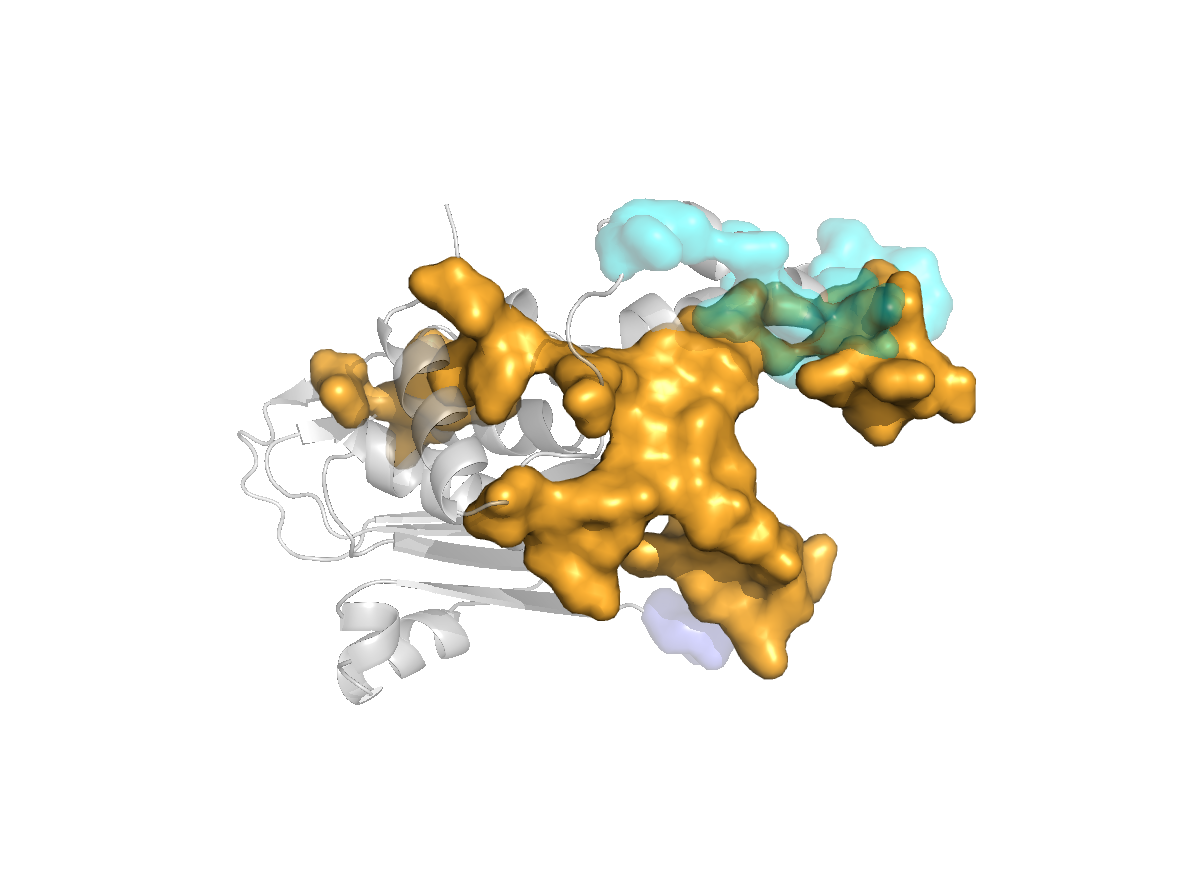 site |
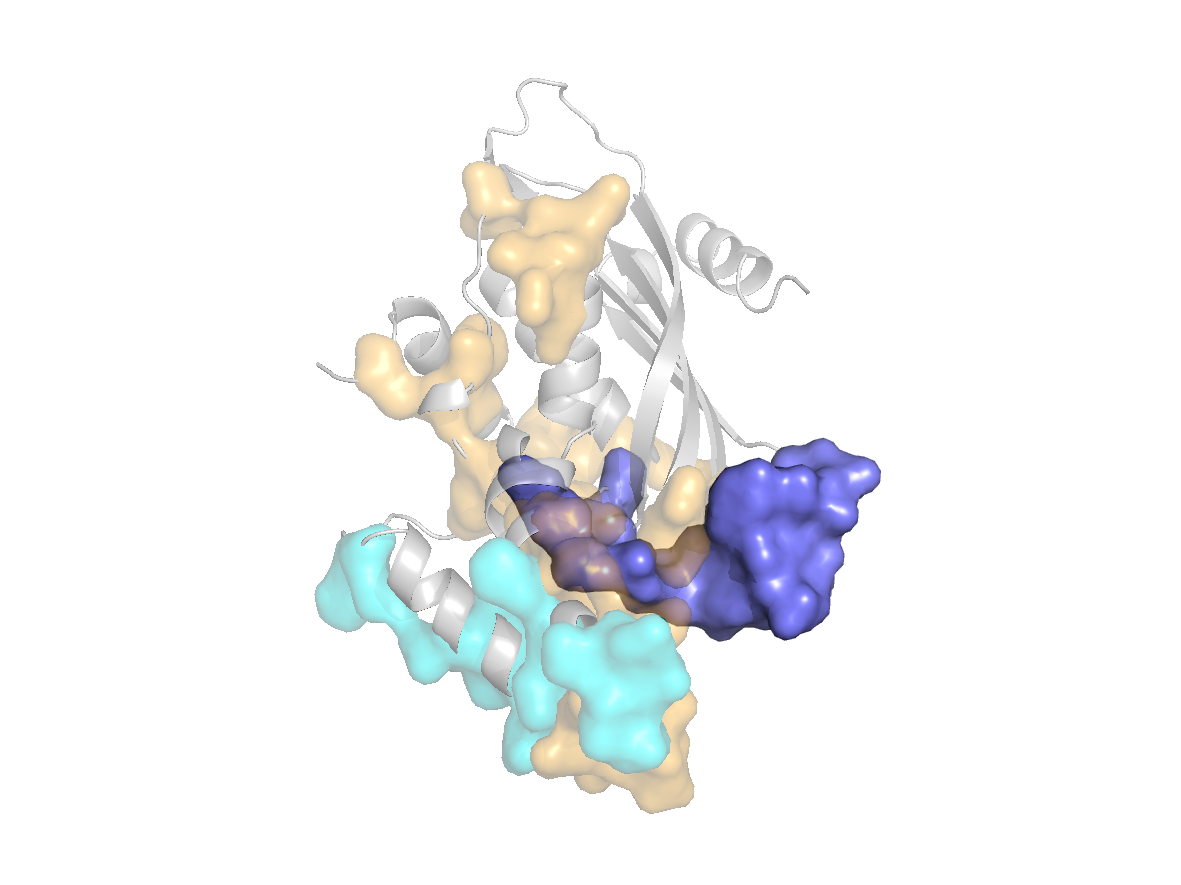 site |
 site |
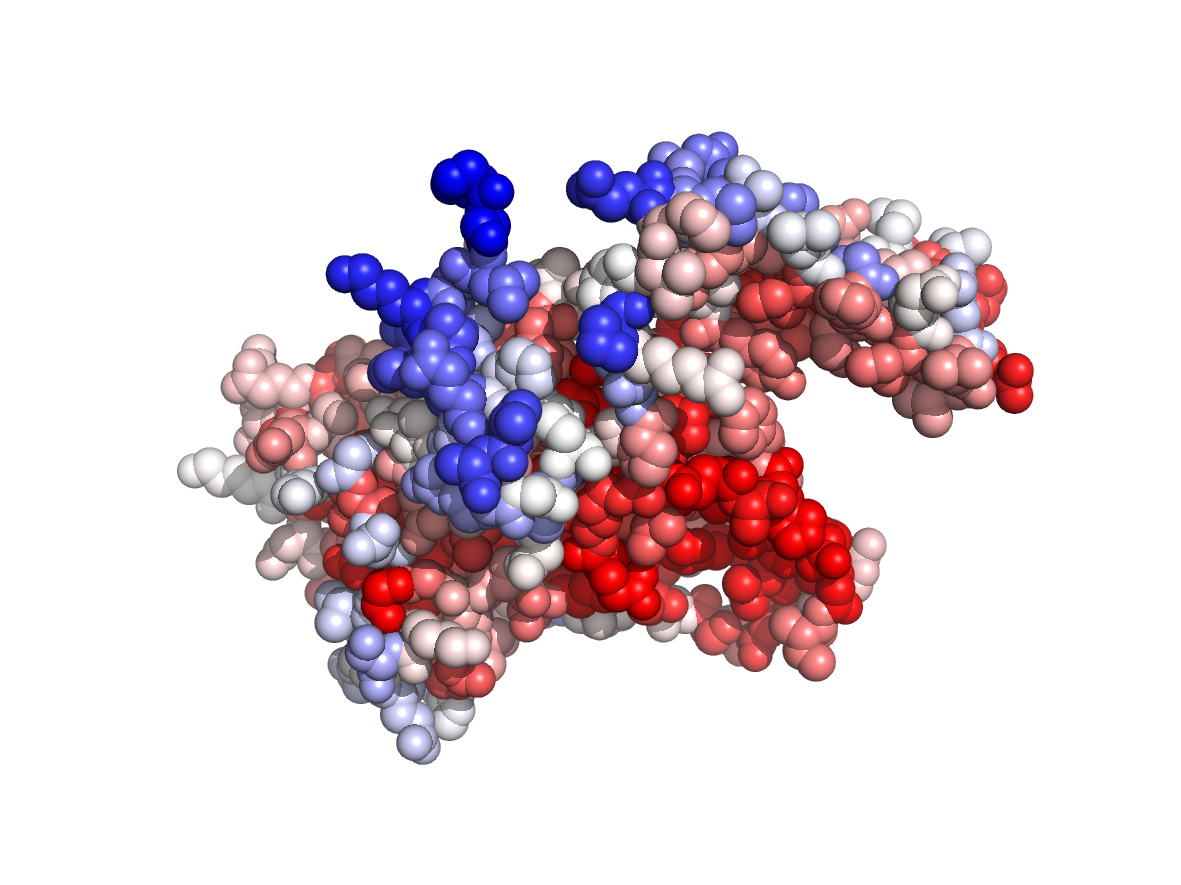
Evolutionary conservation - TJET |
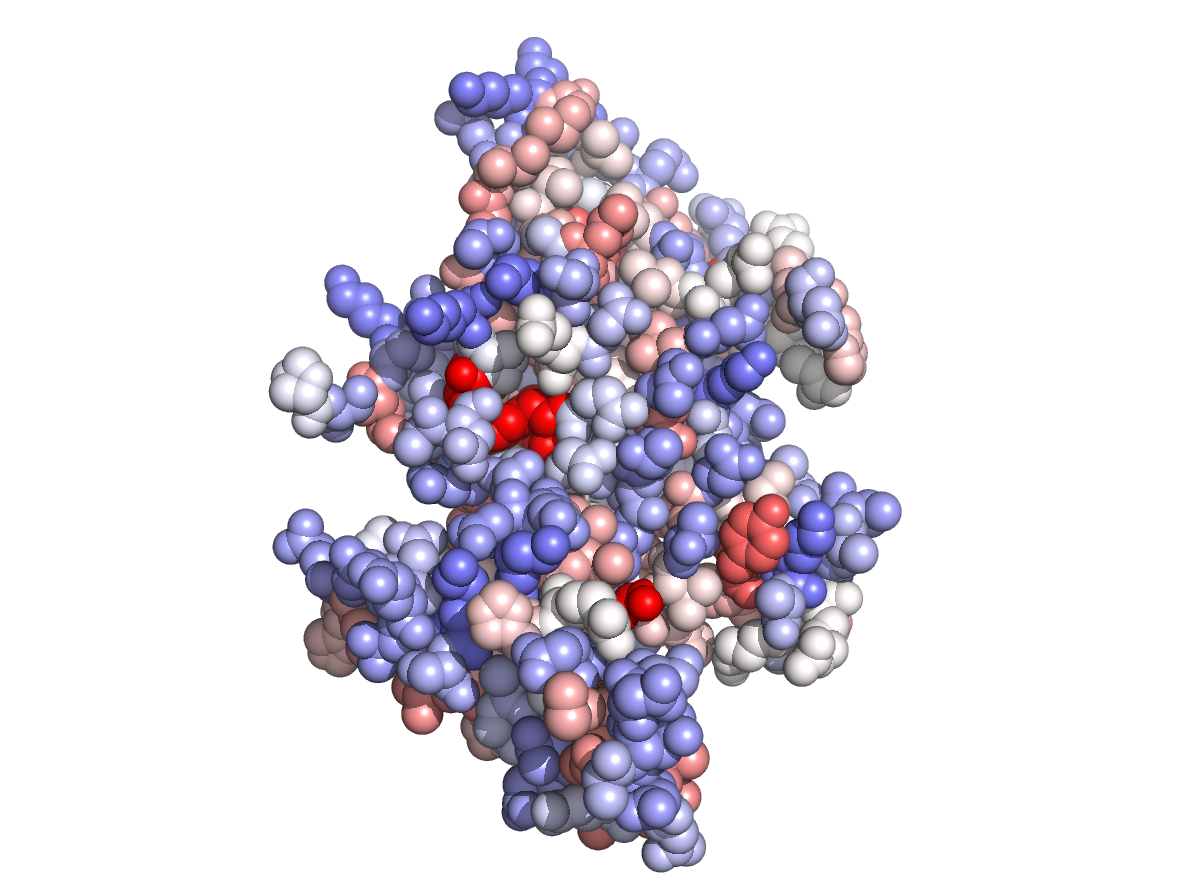
Physico-chemical properties - PC |
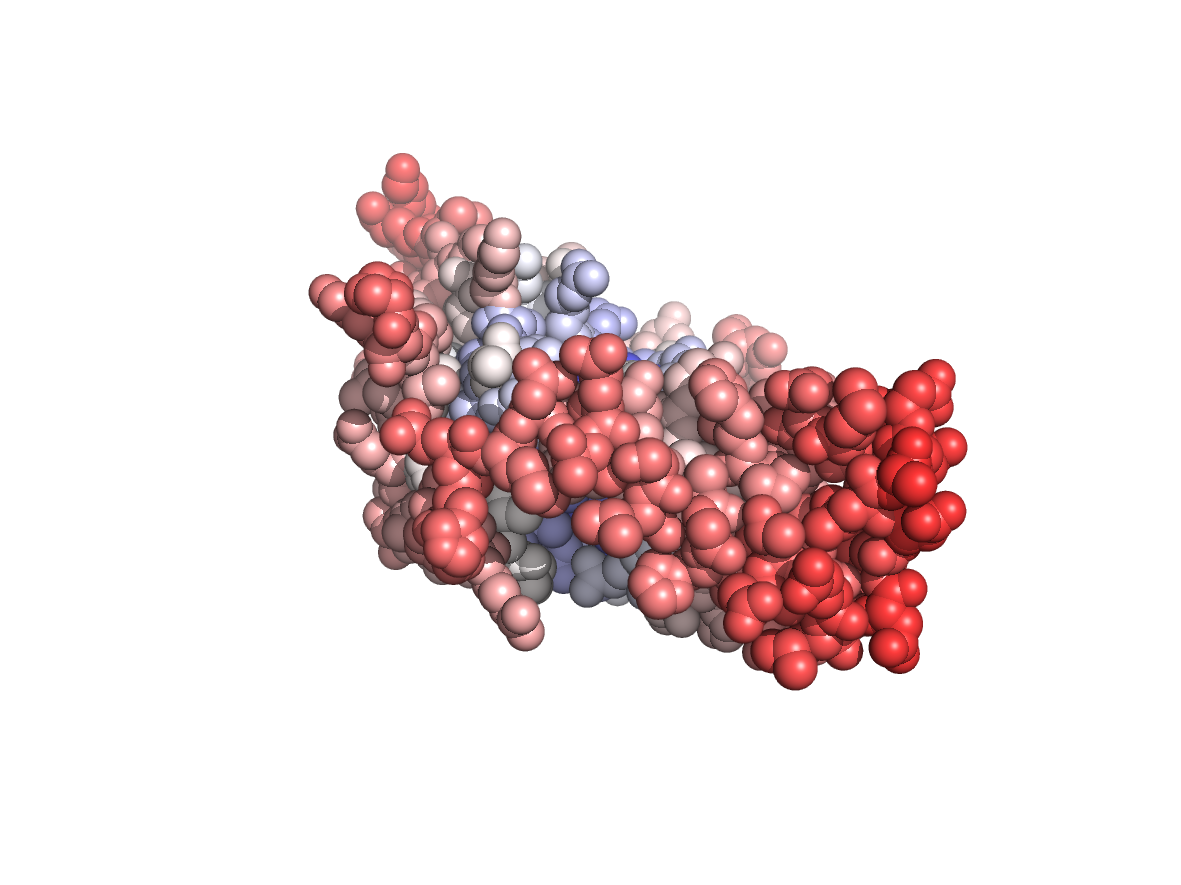
Circular variance - CV |
 |
 |
 |
PDB Structure: 1A8R chain I
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
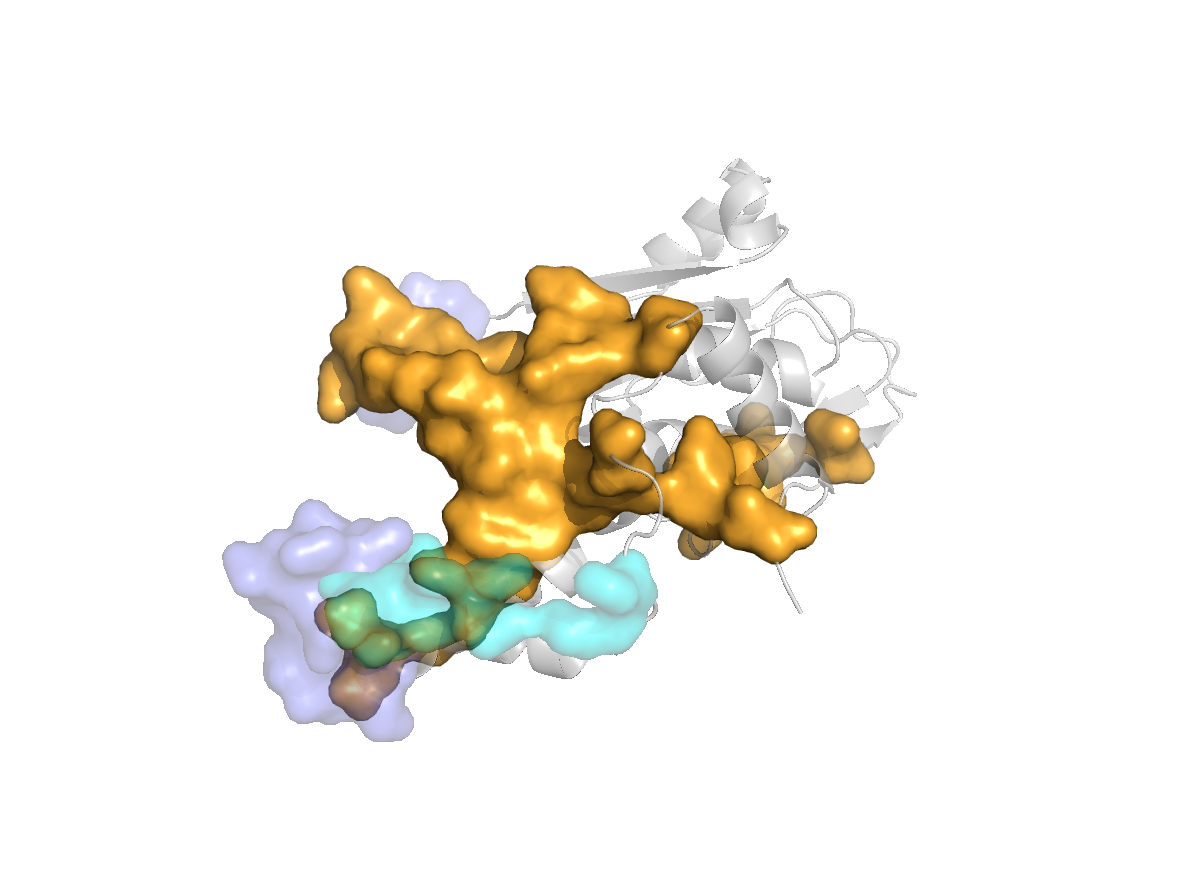 site |
 site |
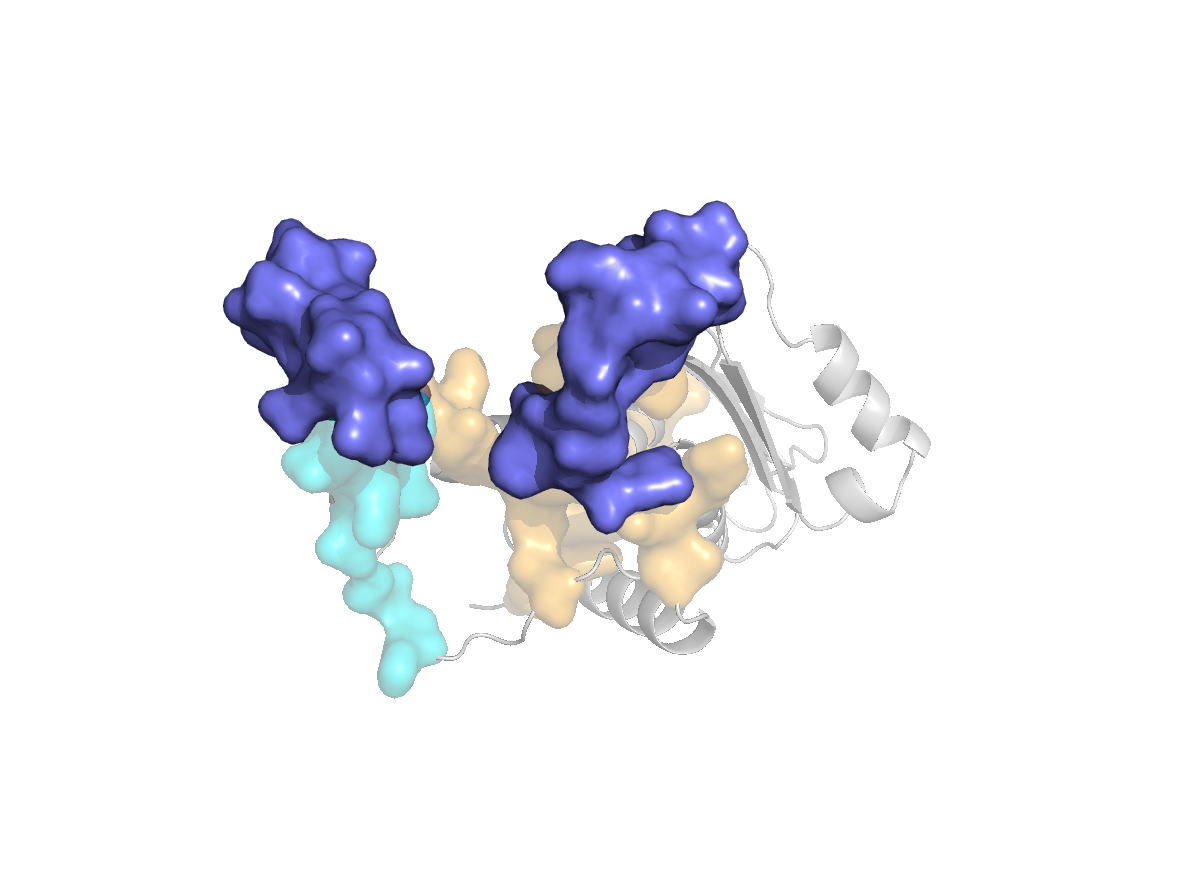
 site |
Evolutionary conservation - TJET |
Physico-chemical properties - PC |
Circular variance - CV |
 |
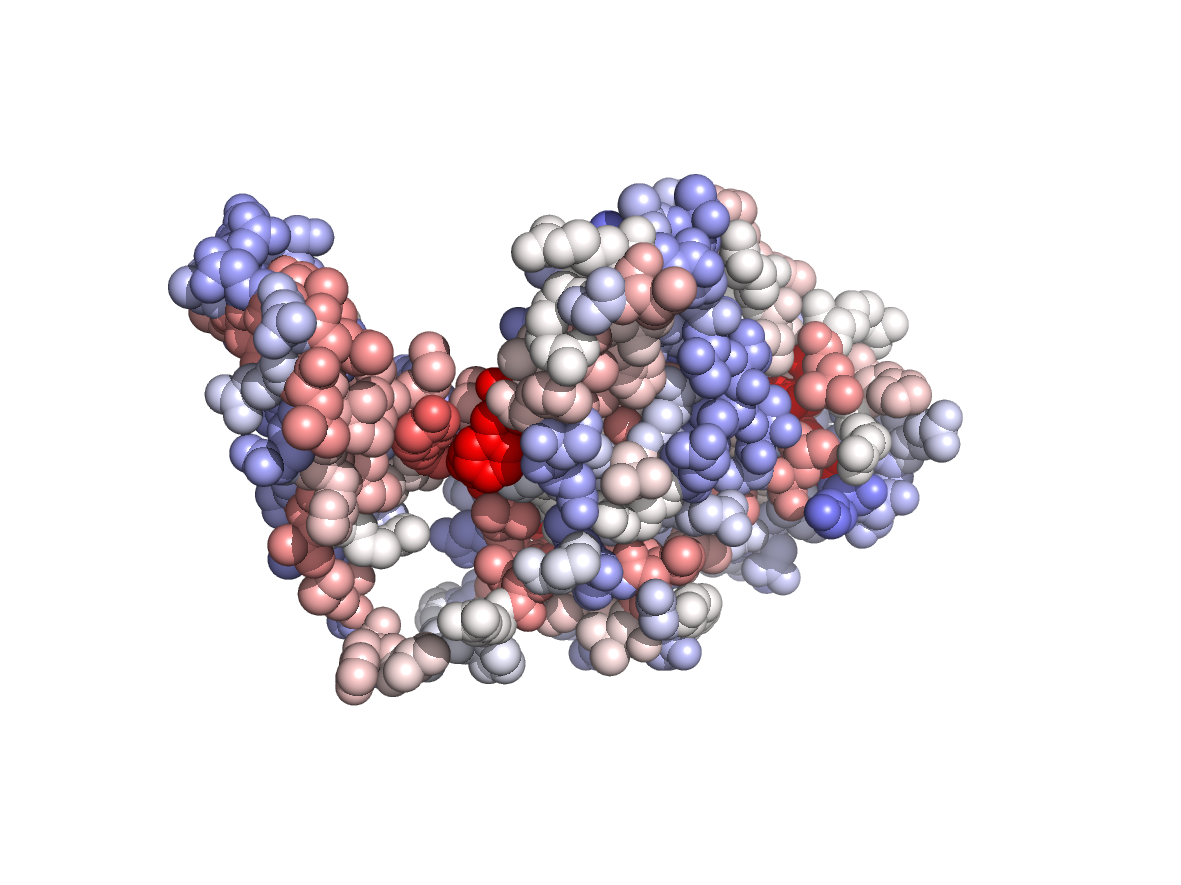 |
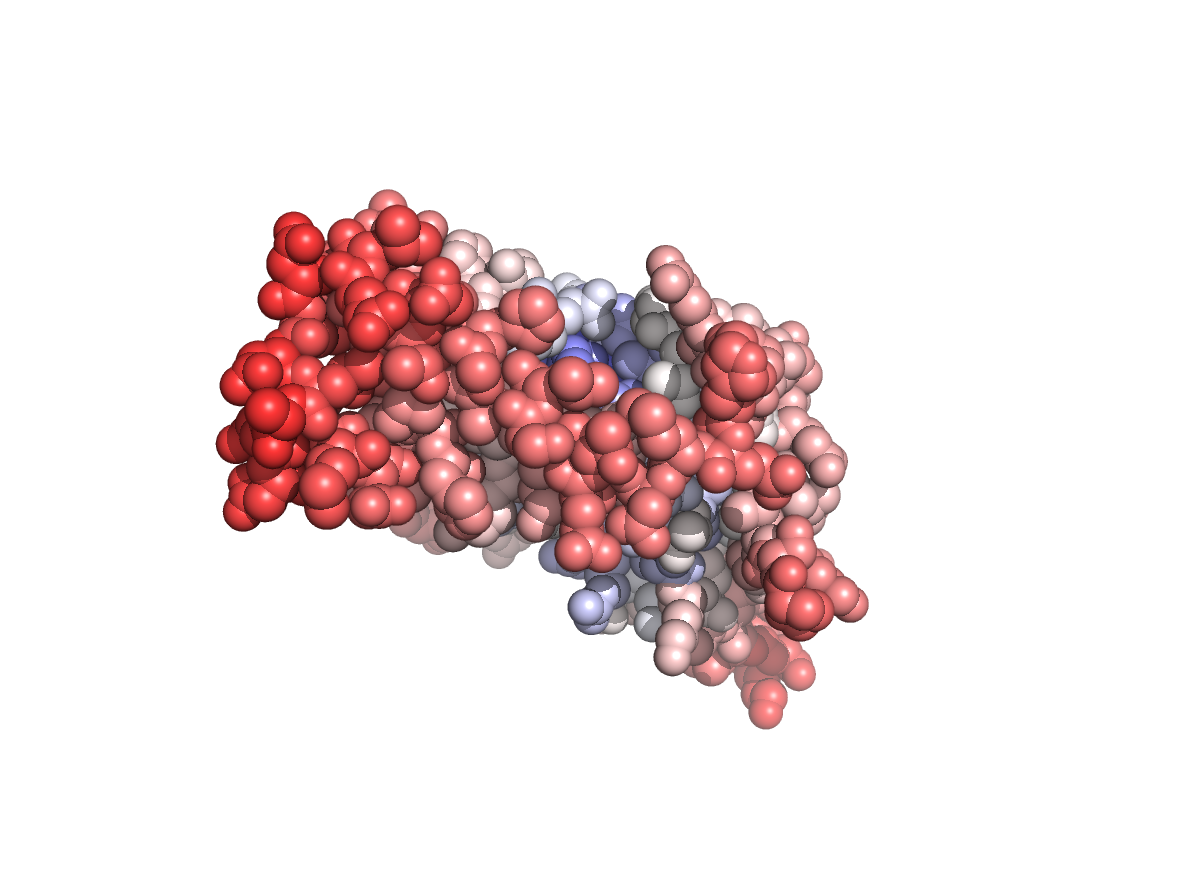 |
PDB Structure: 1A8R chain J
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
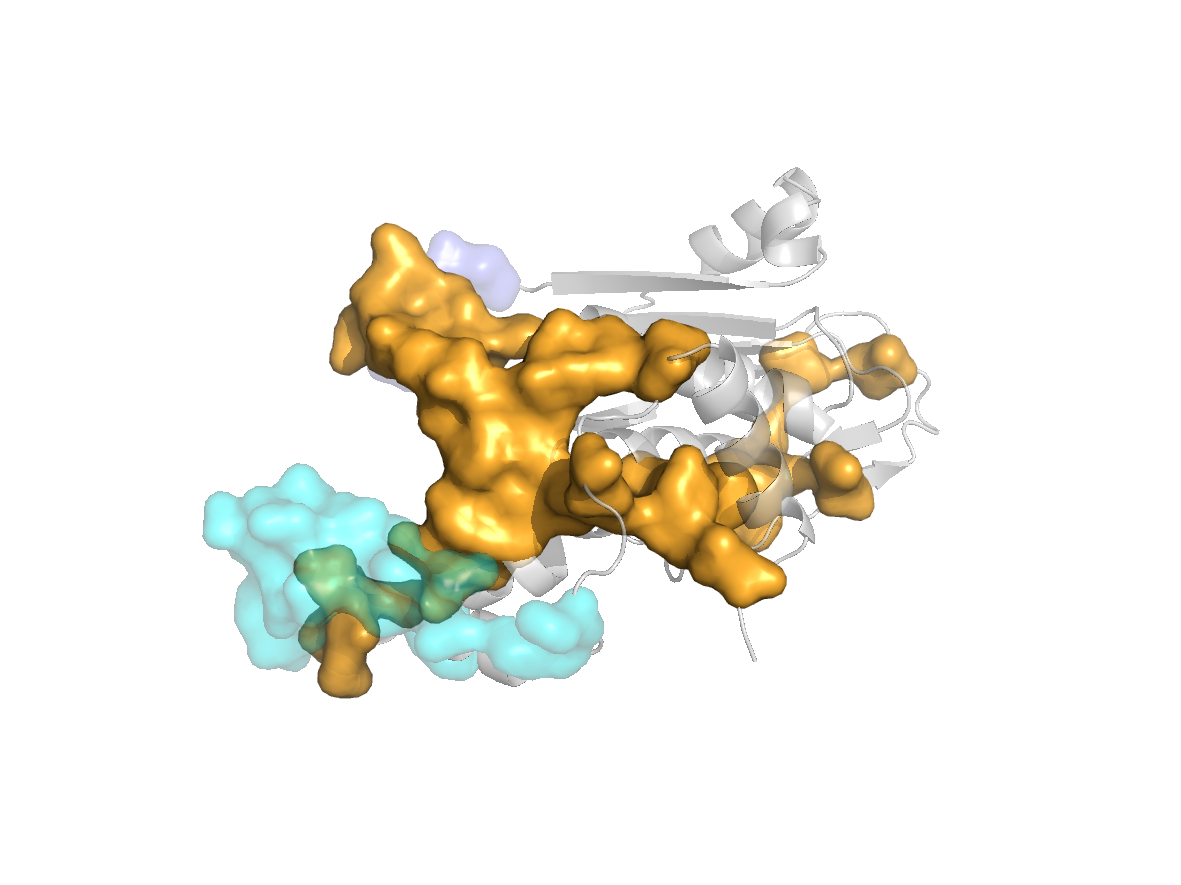 site |
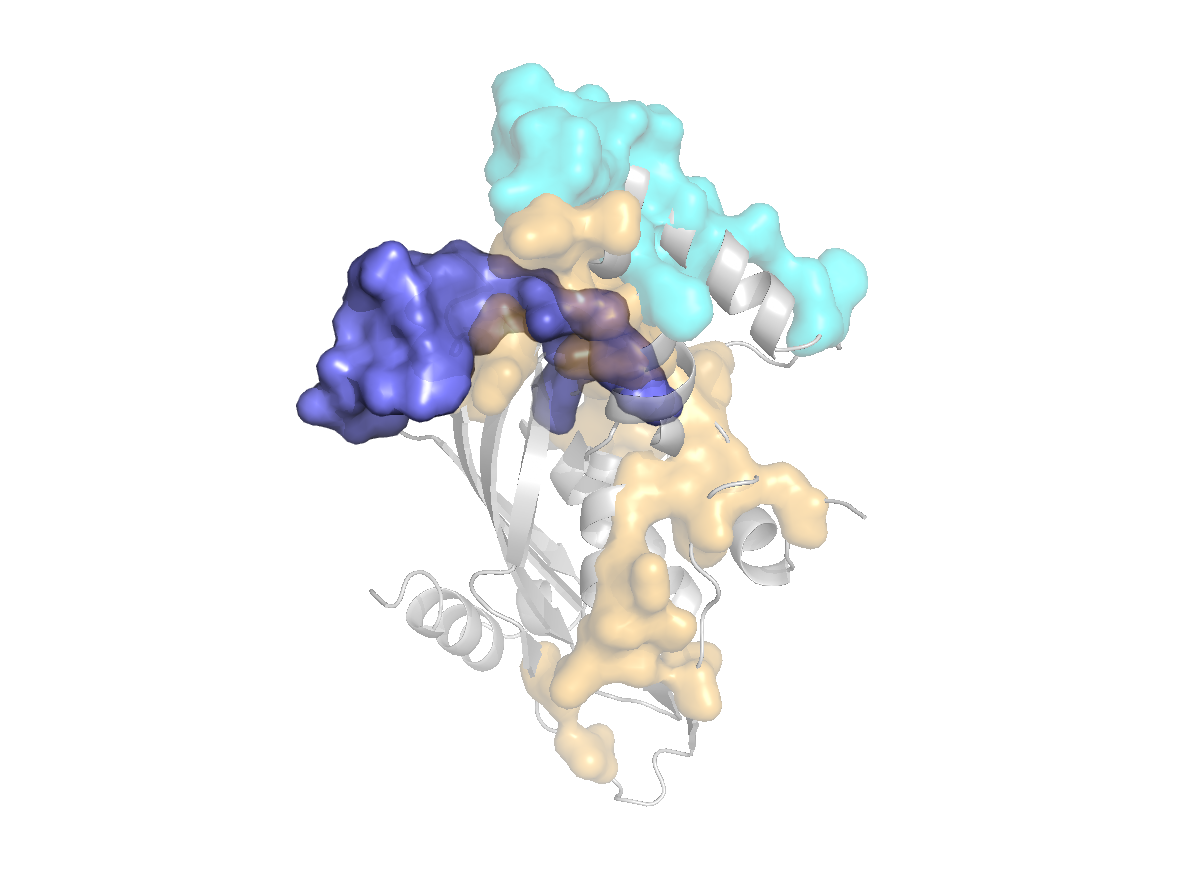 site |
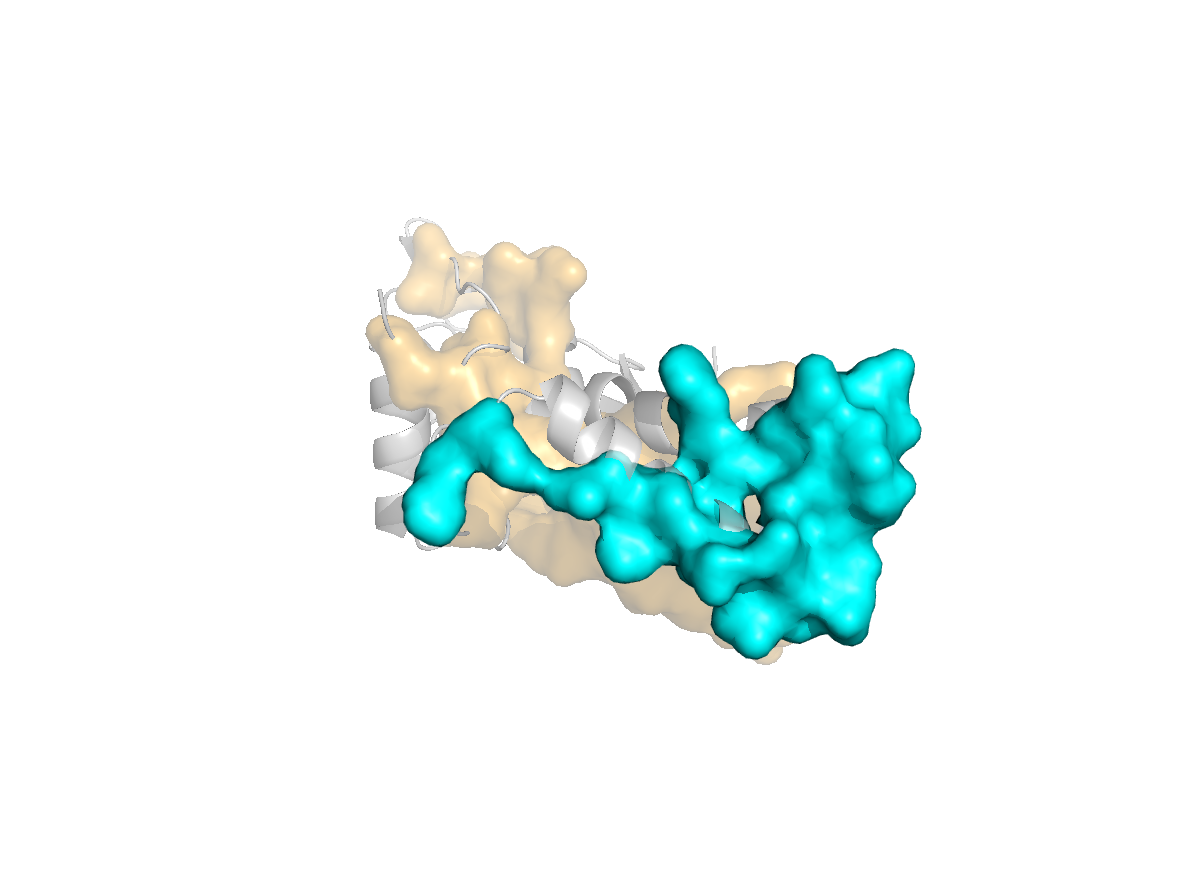 site |
Evolutionary conservation - TJET |
Physico-chemical properties - PC |
Circular variance - CV |
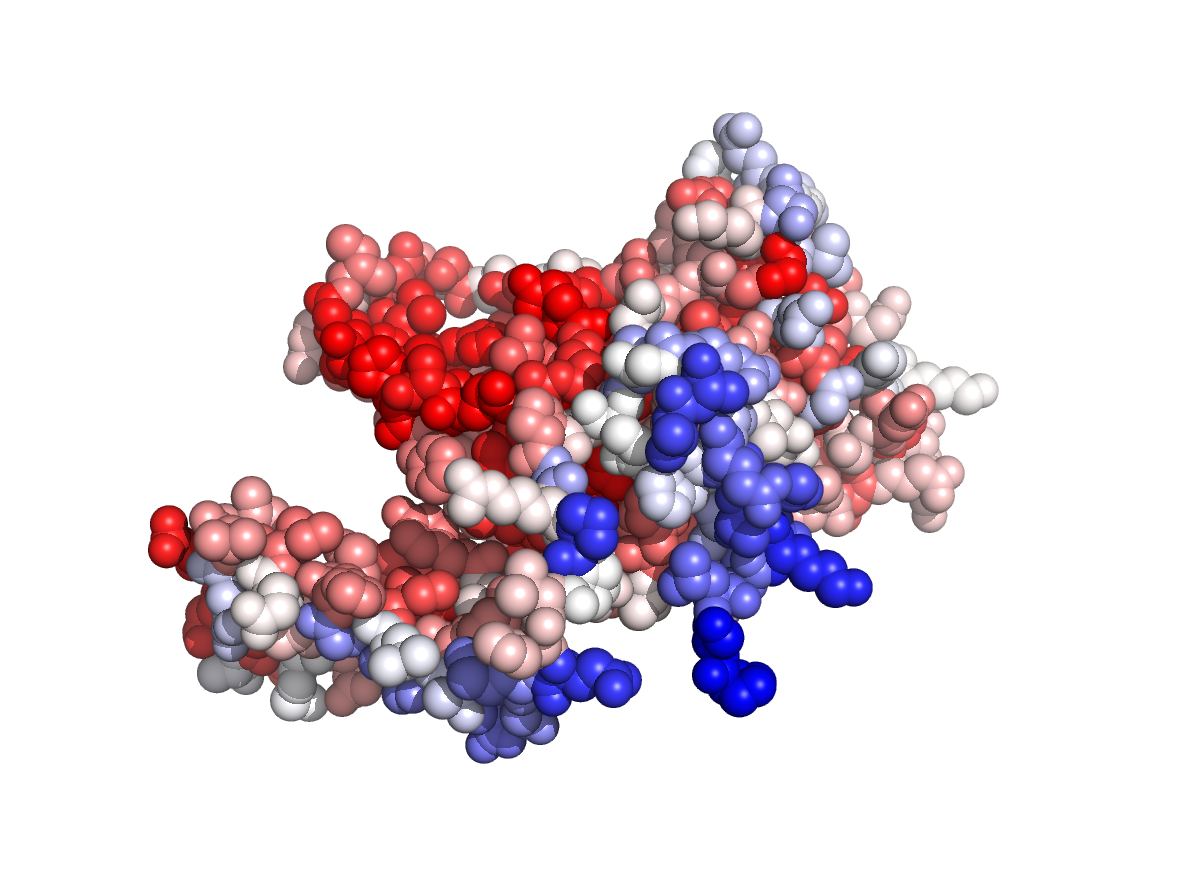 |
 |
 |
PDB Structure: 1A8R chain K
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
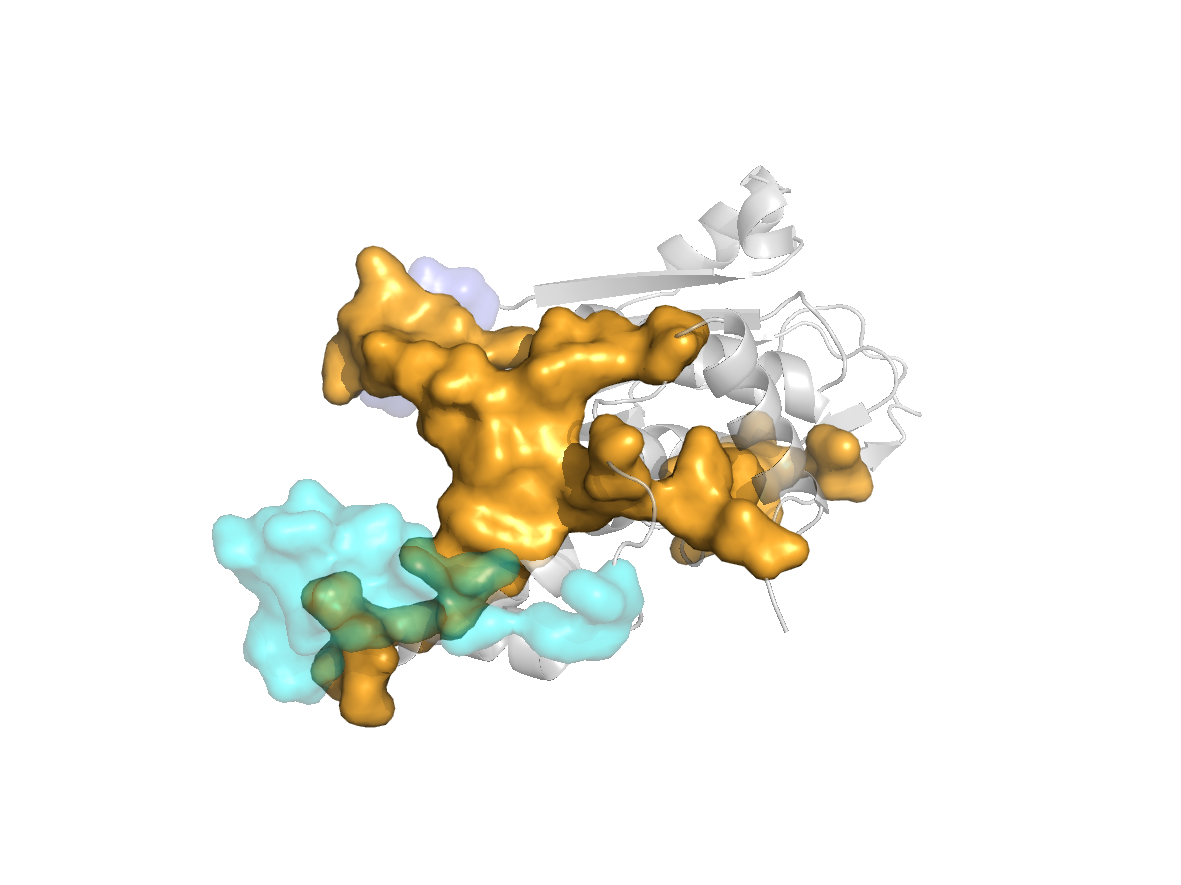 site |
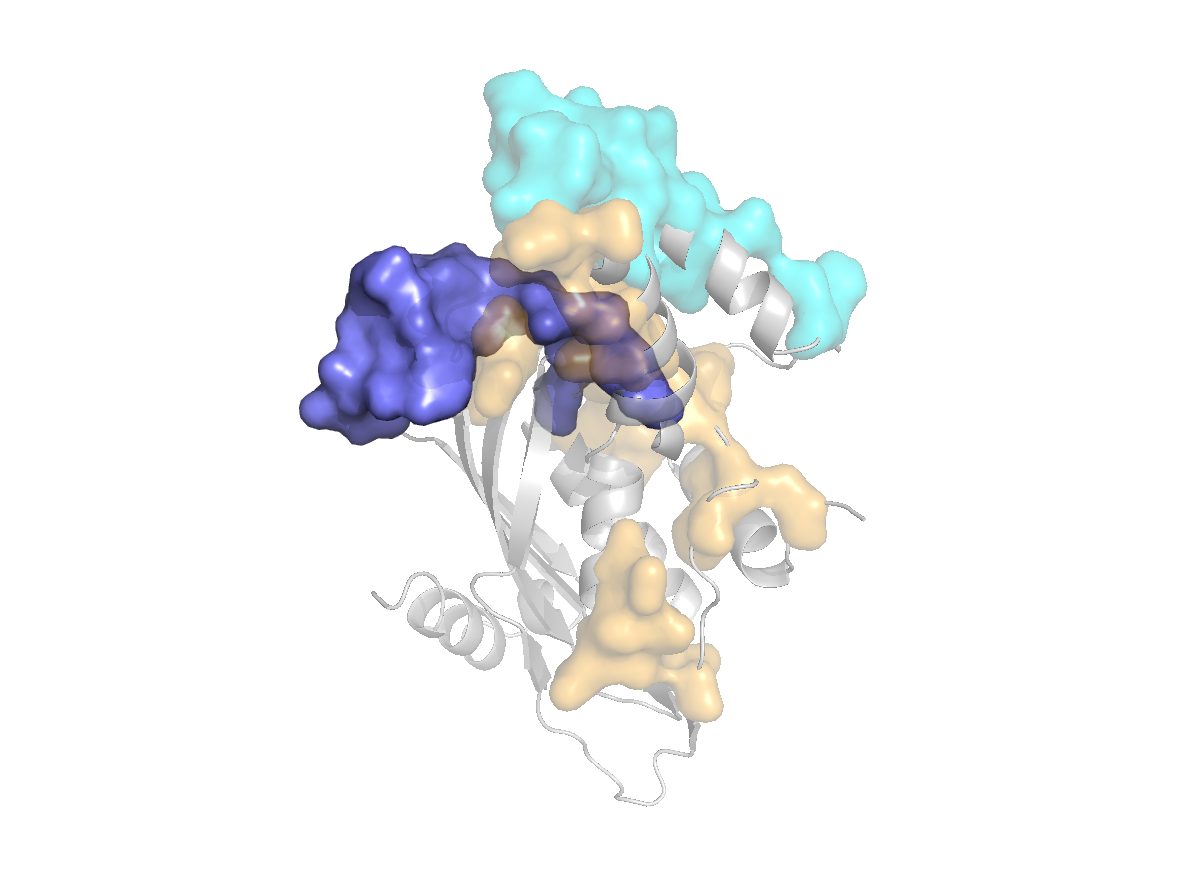 site |
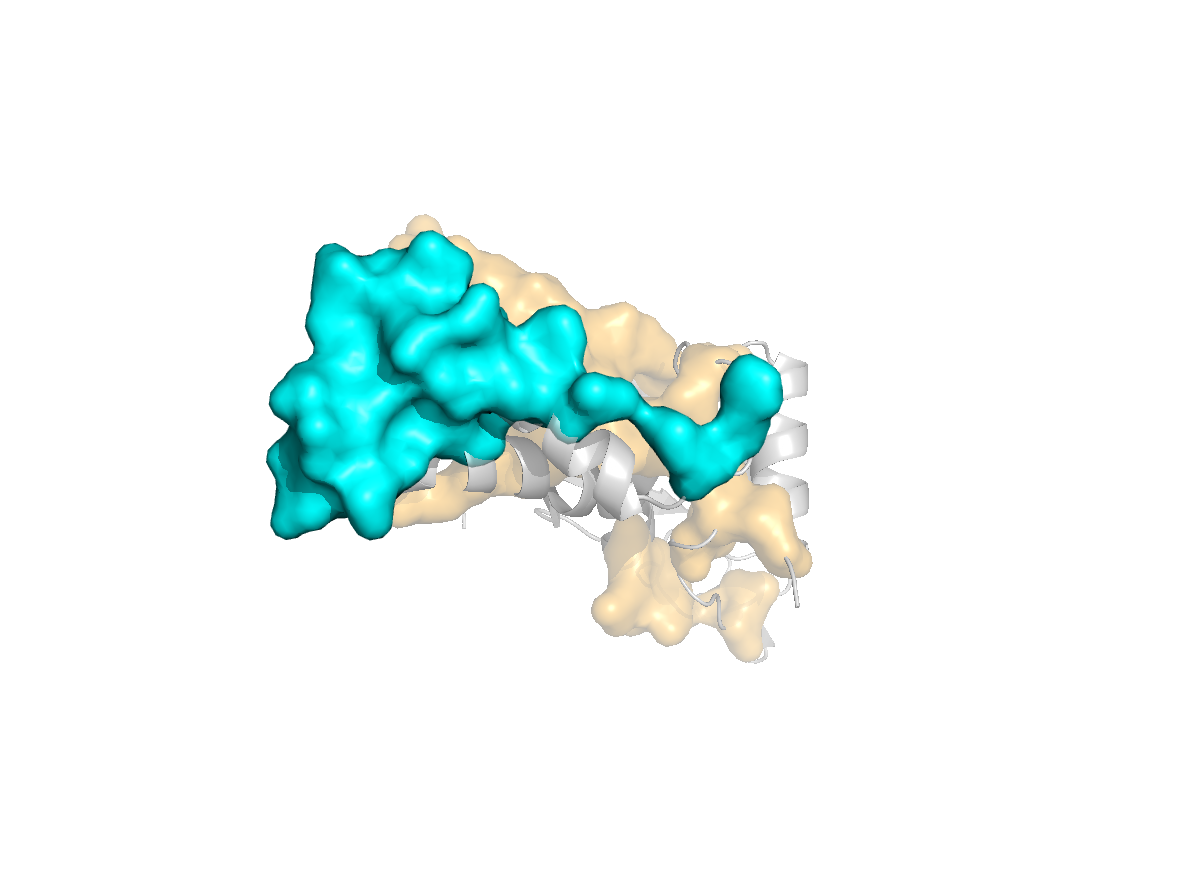 site |
Evolutionary conservation - TJET |
Physico-chemical properties - PC |
Circular variance - CV |
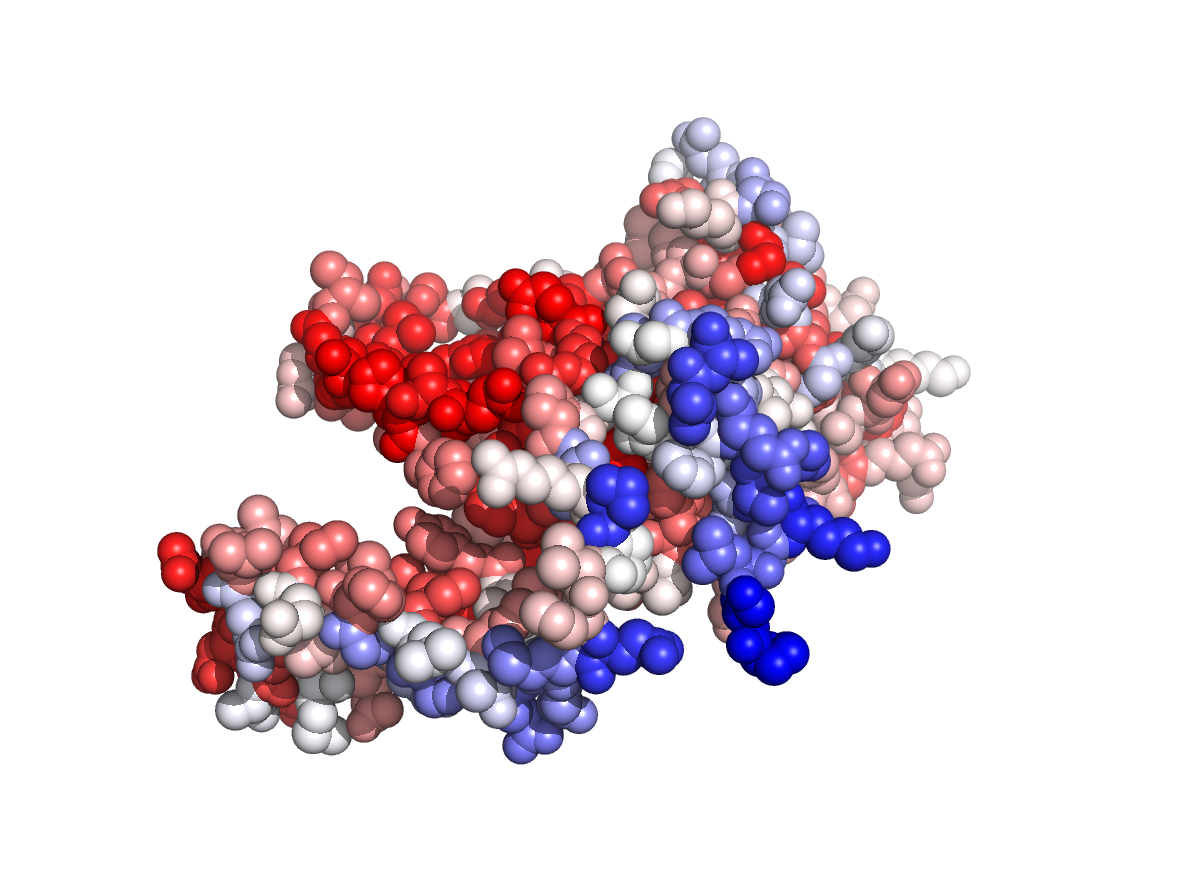 |
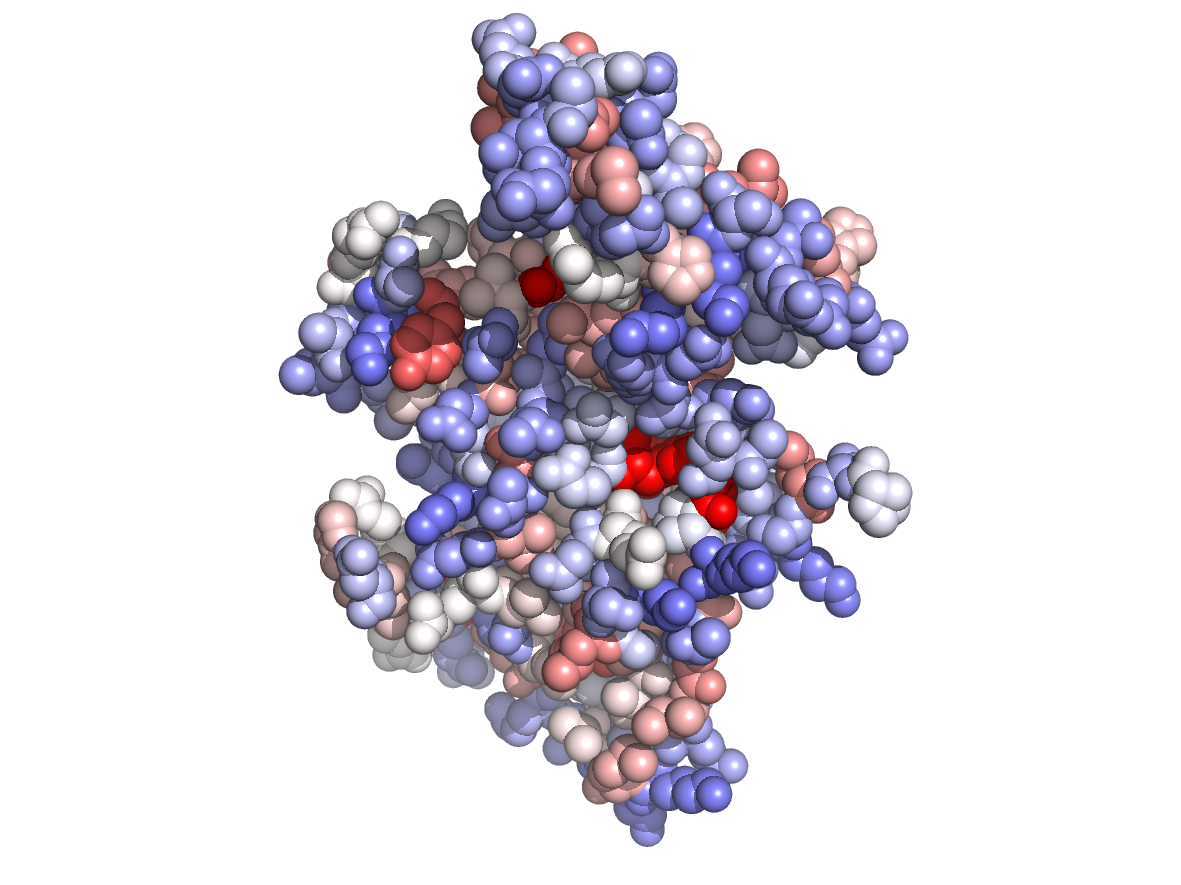 |
 |
PDB Structure: 1A8R chain L
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
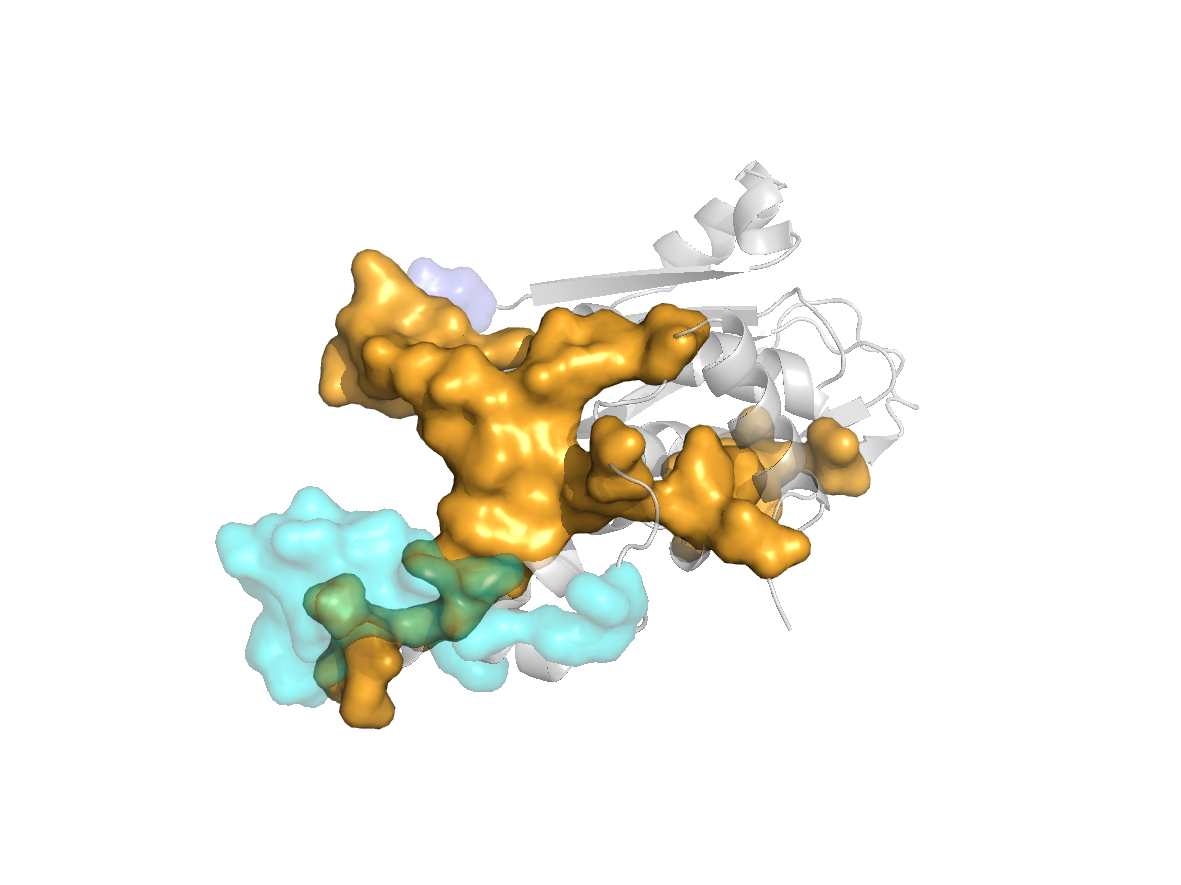 site |
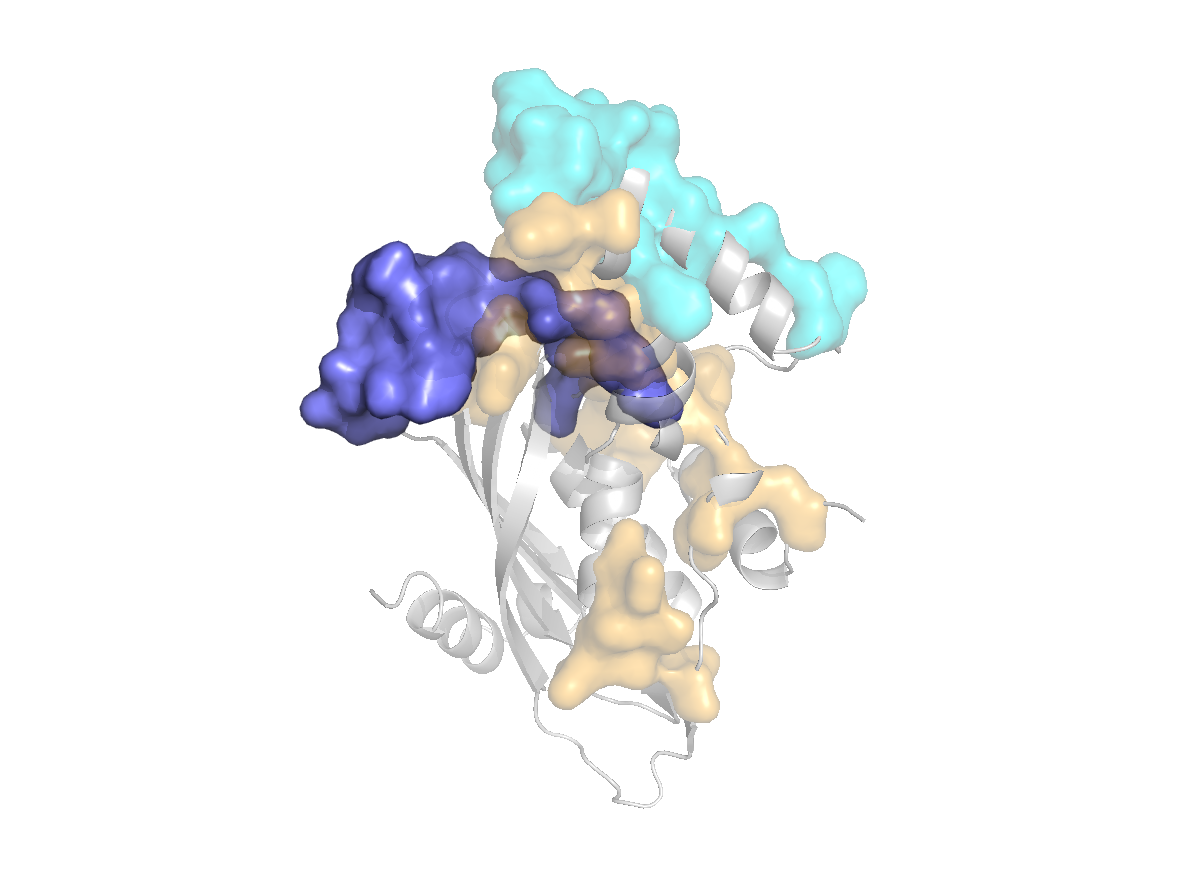 site |
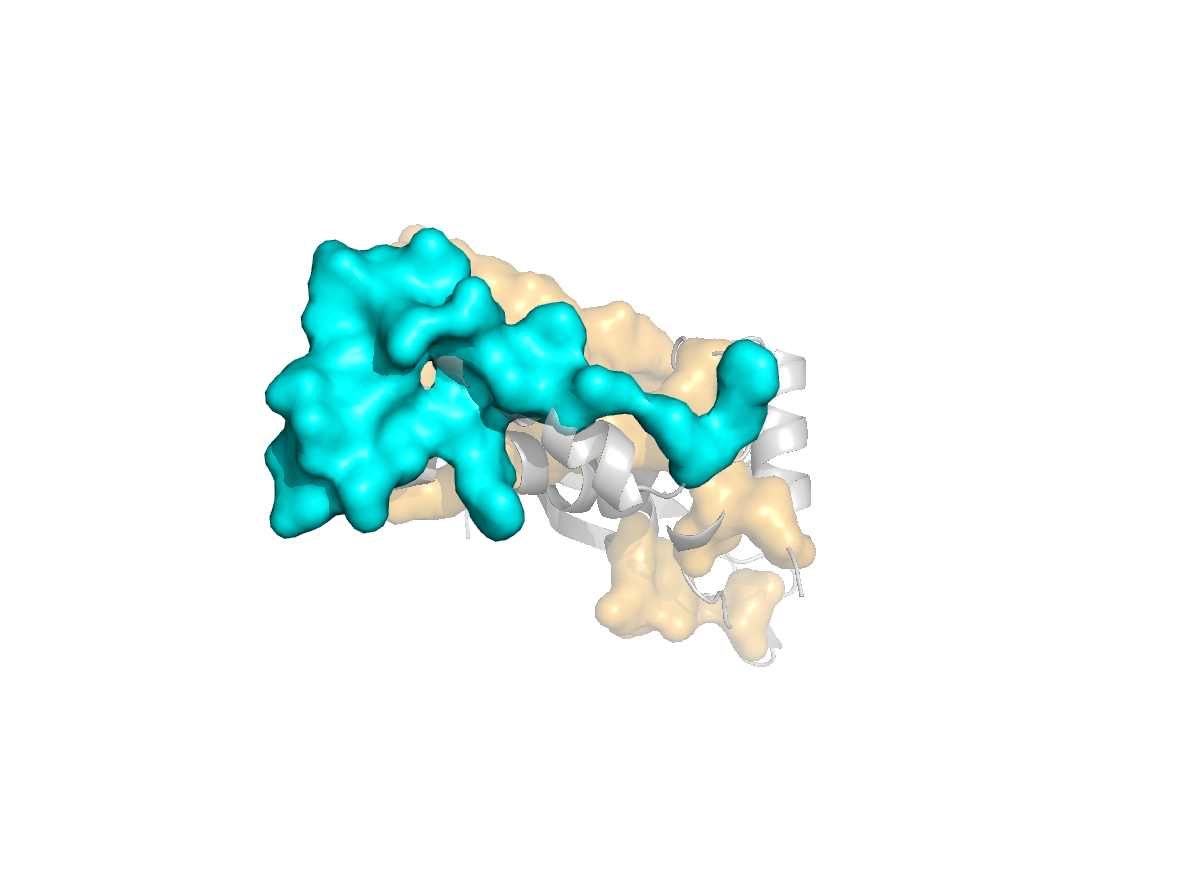 site |
Evolutionary conservation - TJET |
Physico-chemical properties - PC |
Circular variance - CV |
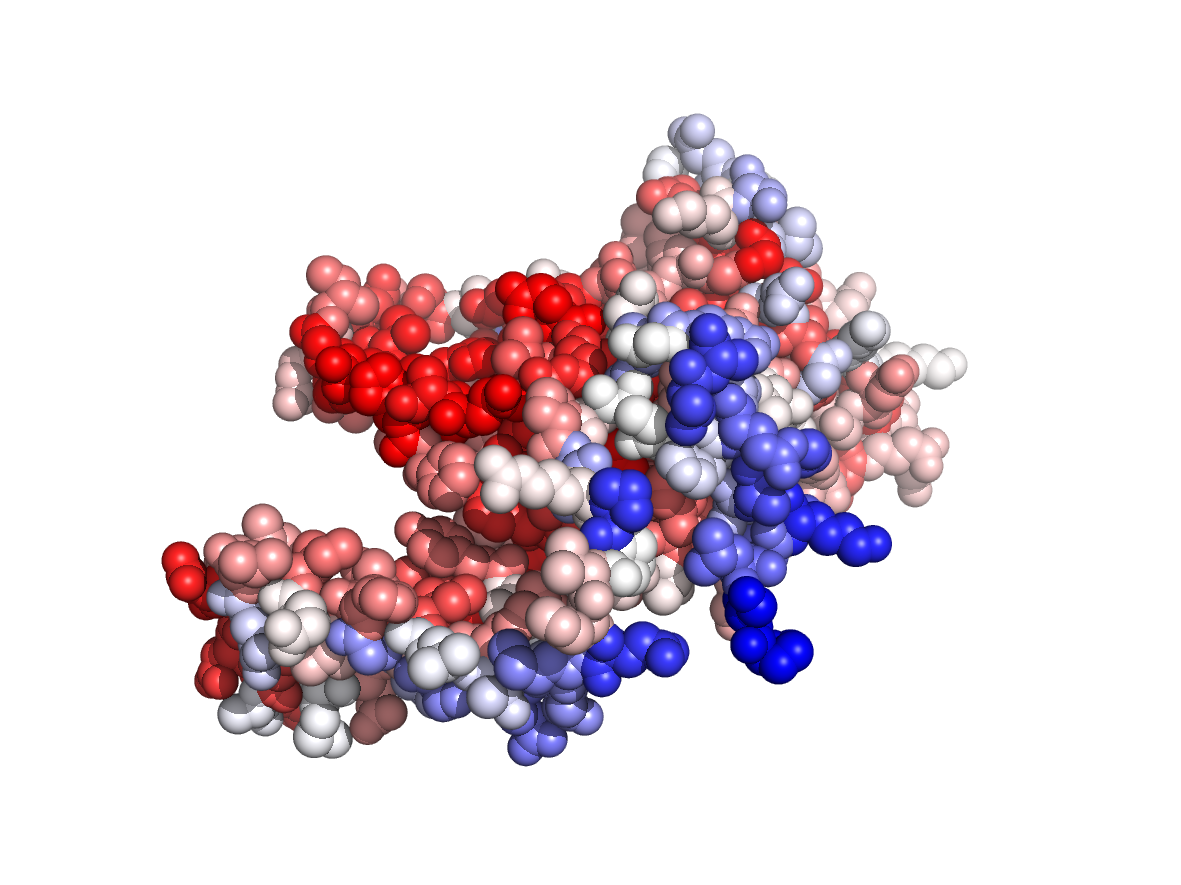 |
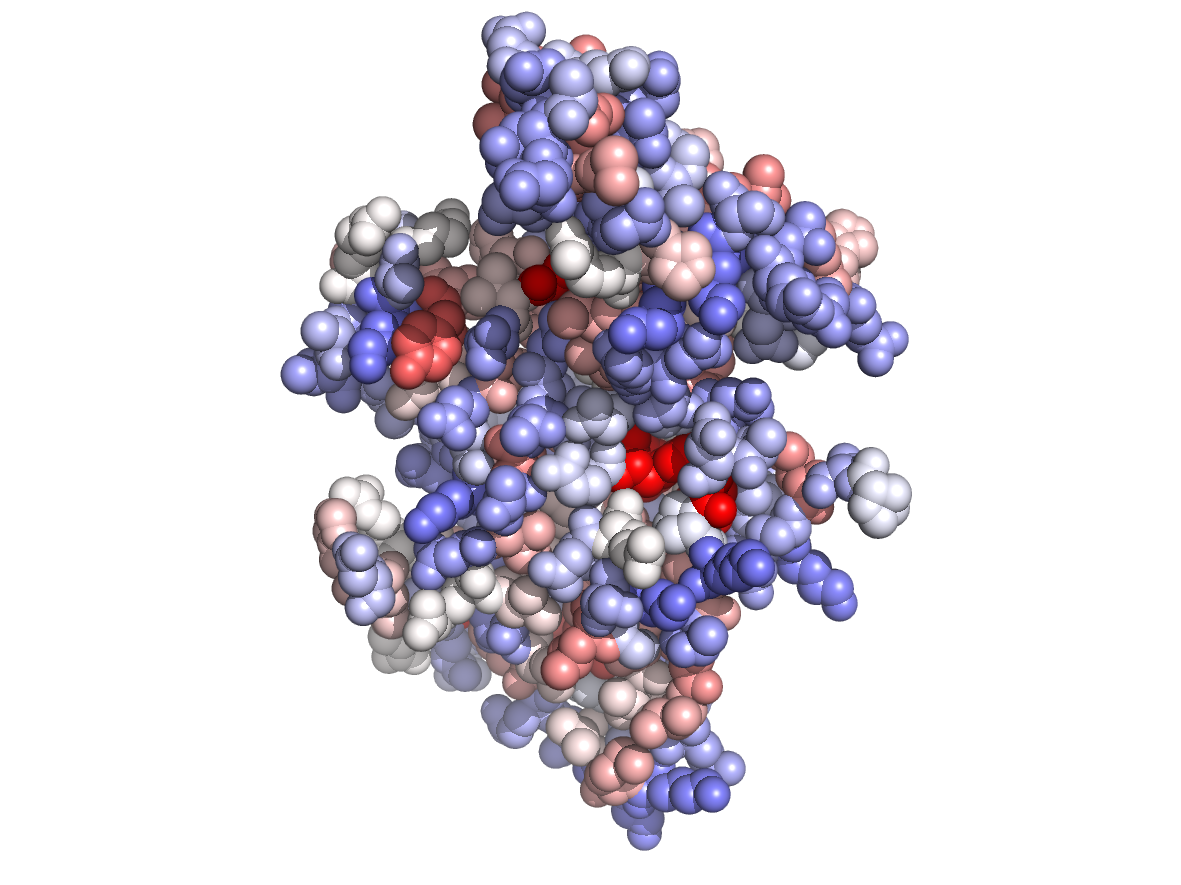 |
 |
PDB Structure: 1A8R chain M
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
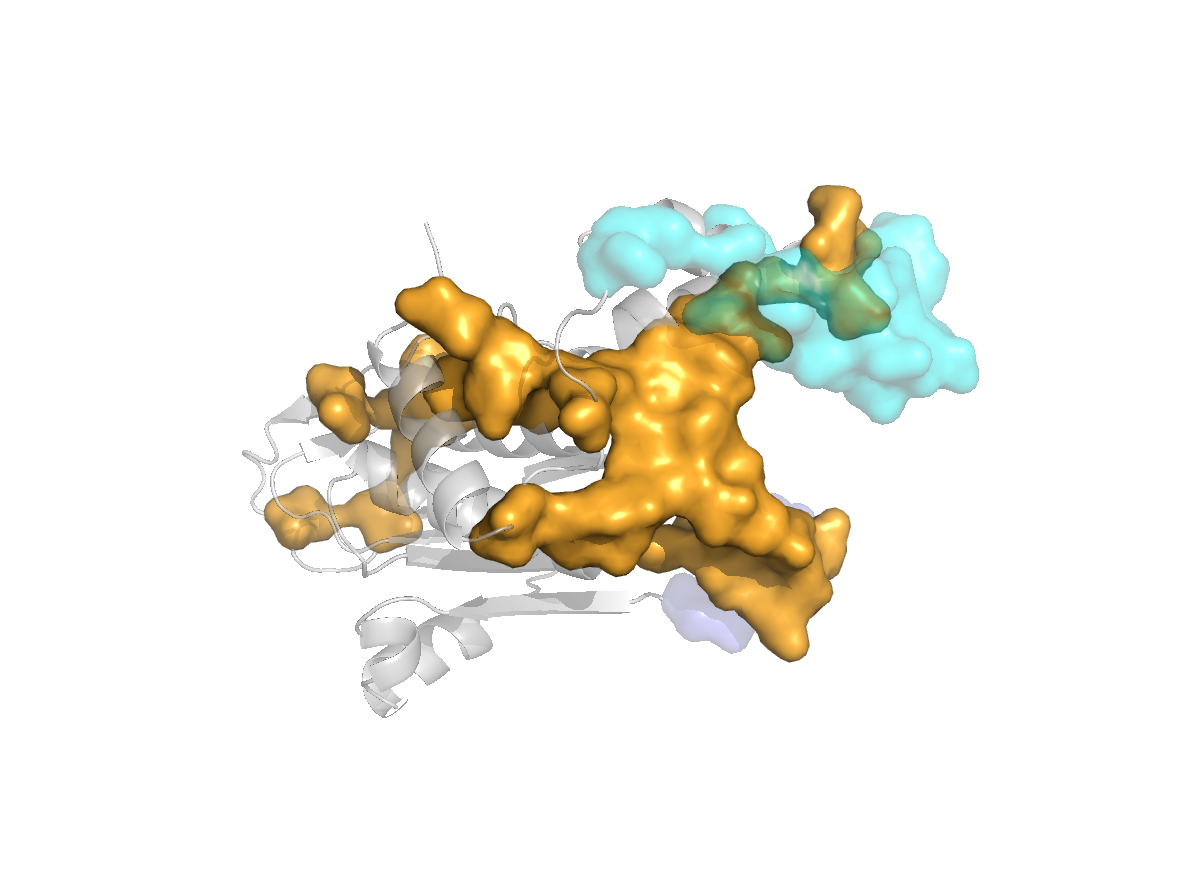 site |
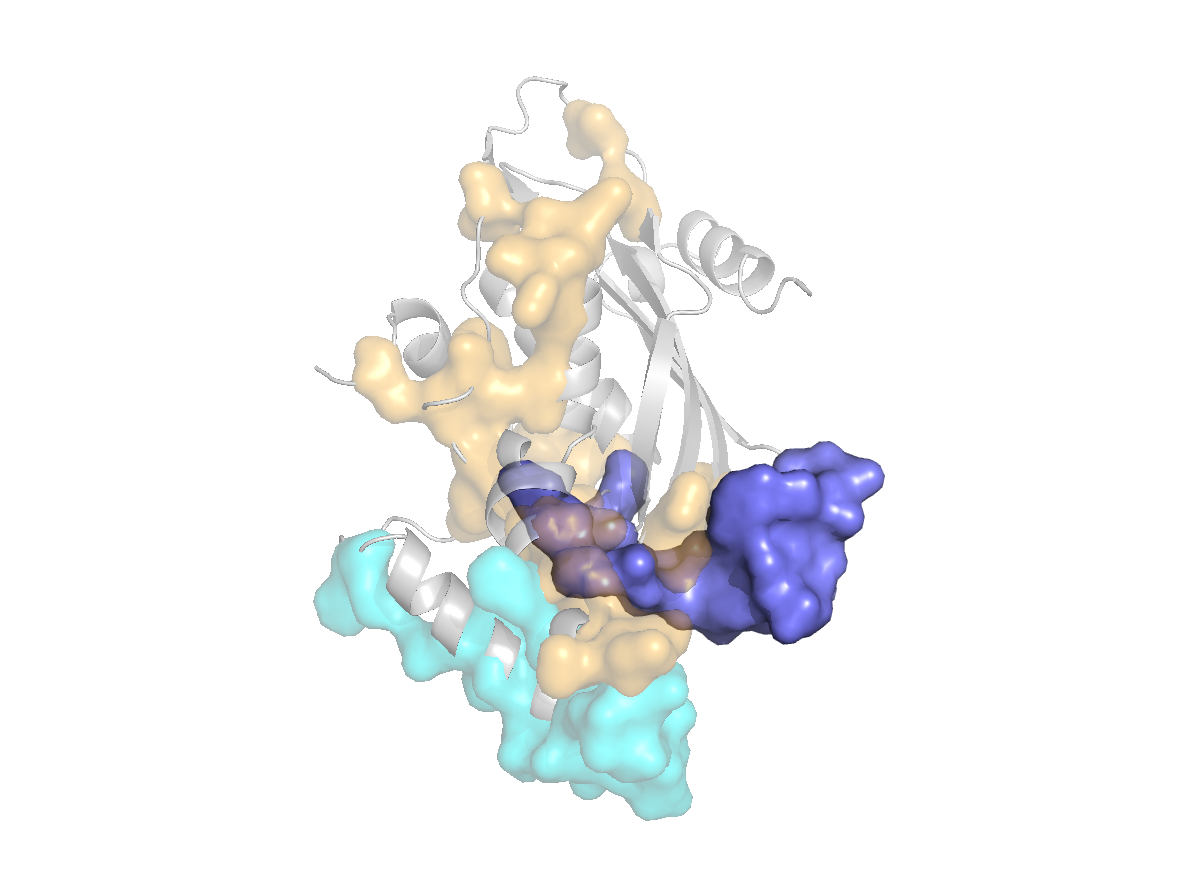 site |
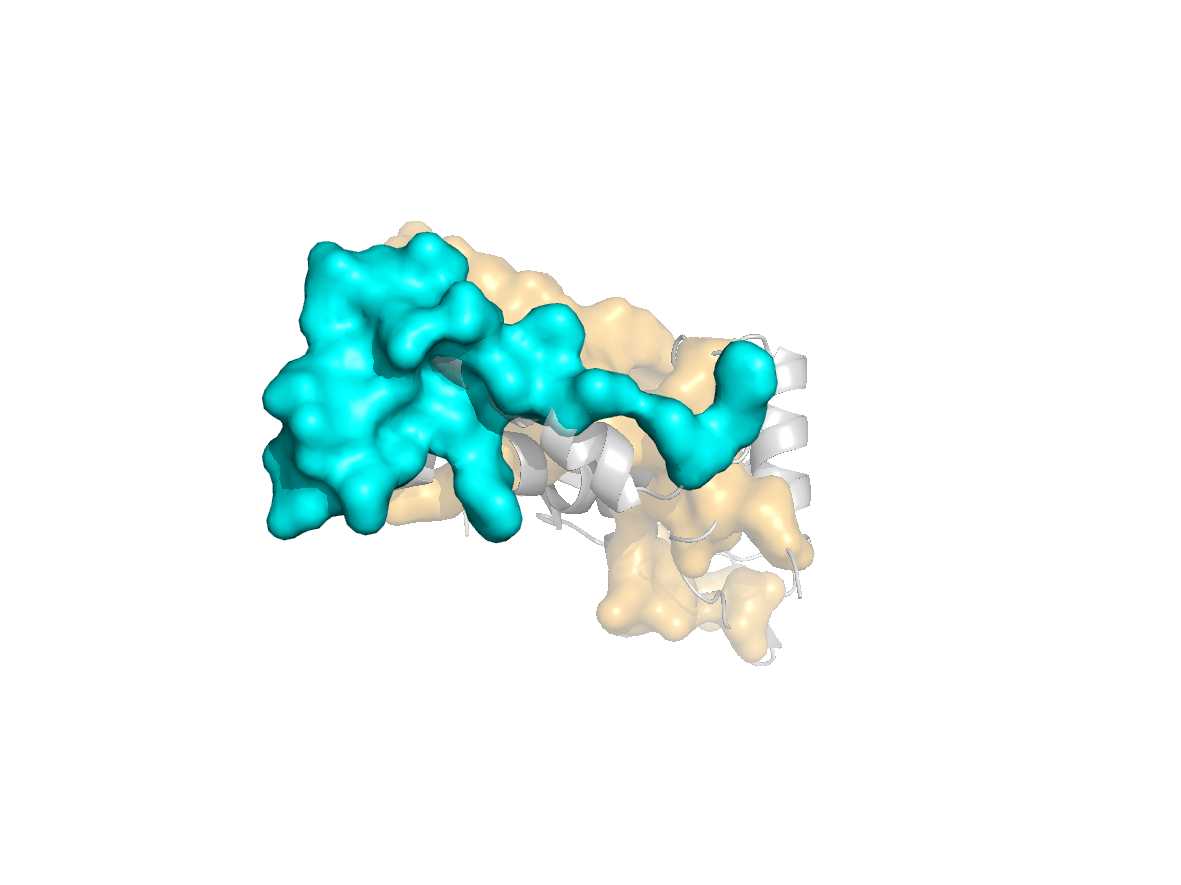 site |
Evolutionary conservation - TJET |
Physico-chemical properties - PC |
Circular variance - CV |
 |
 |
 |
PDB Structure: 1A8R chain N
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
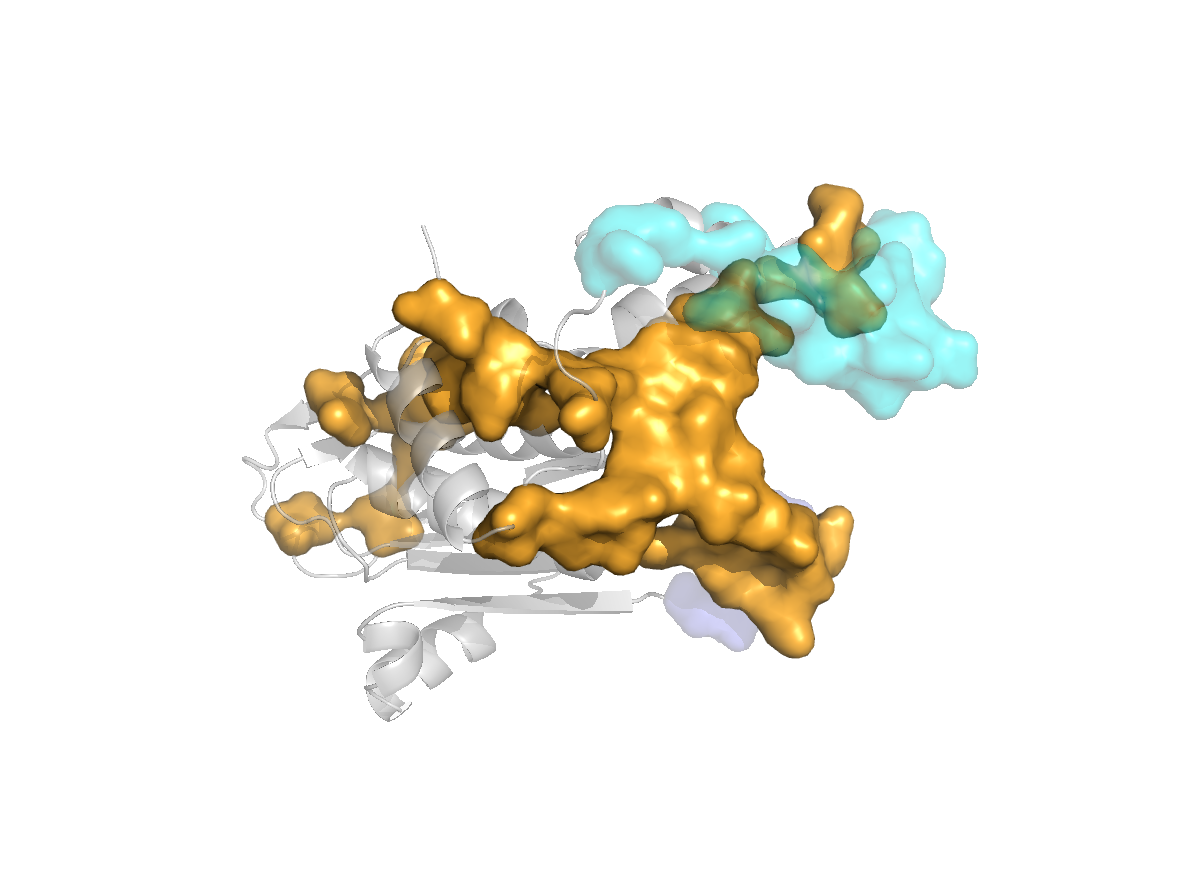 site |
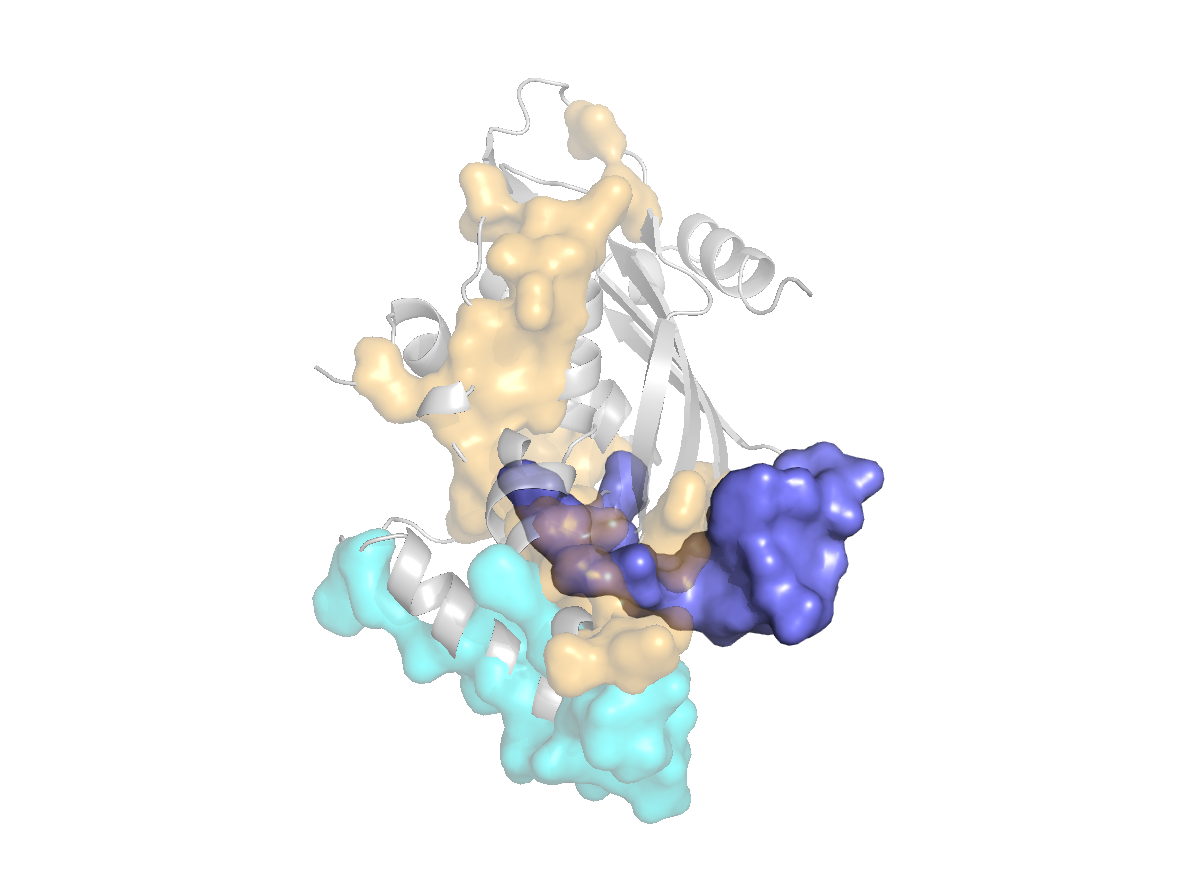 site |
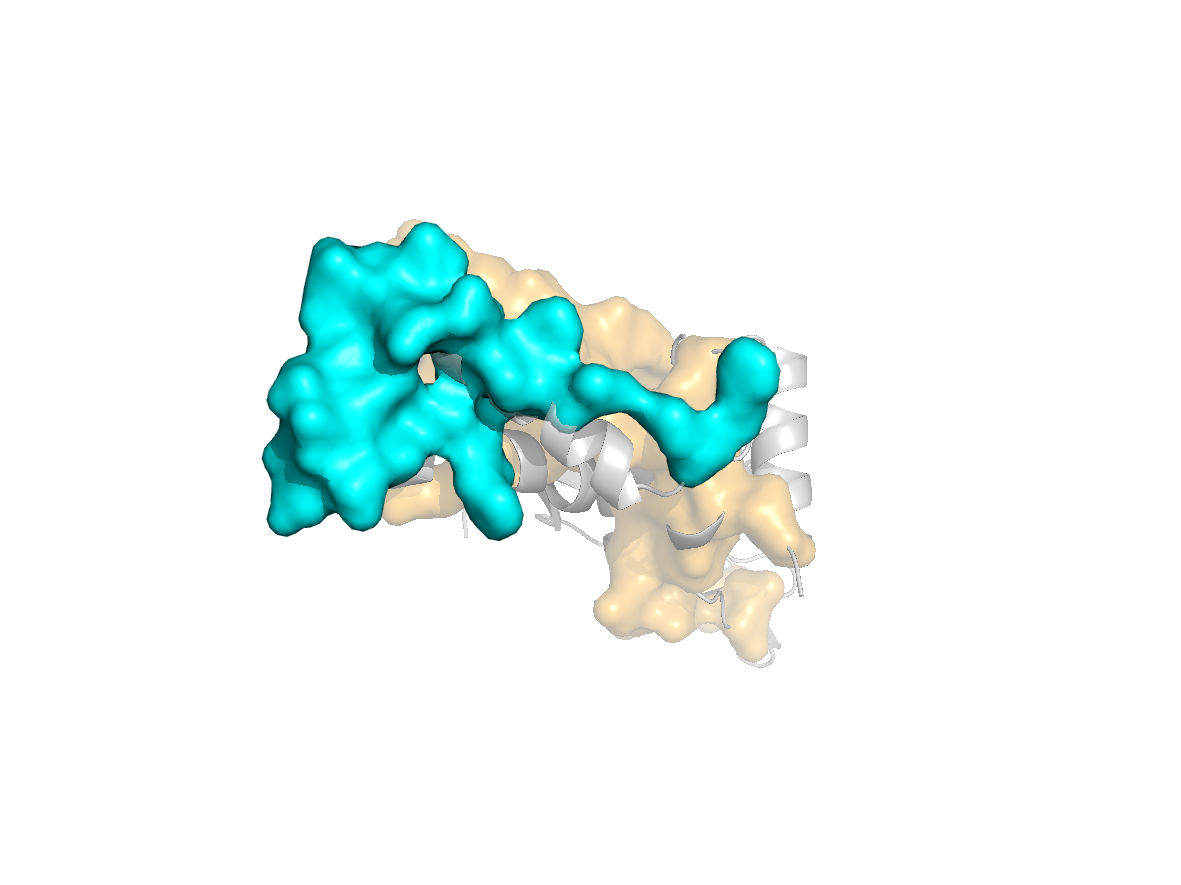 site |
Evolutionary conservation - TJET |
Physico-chemical properties - PC |
Circular variance - CV |
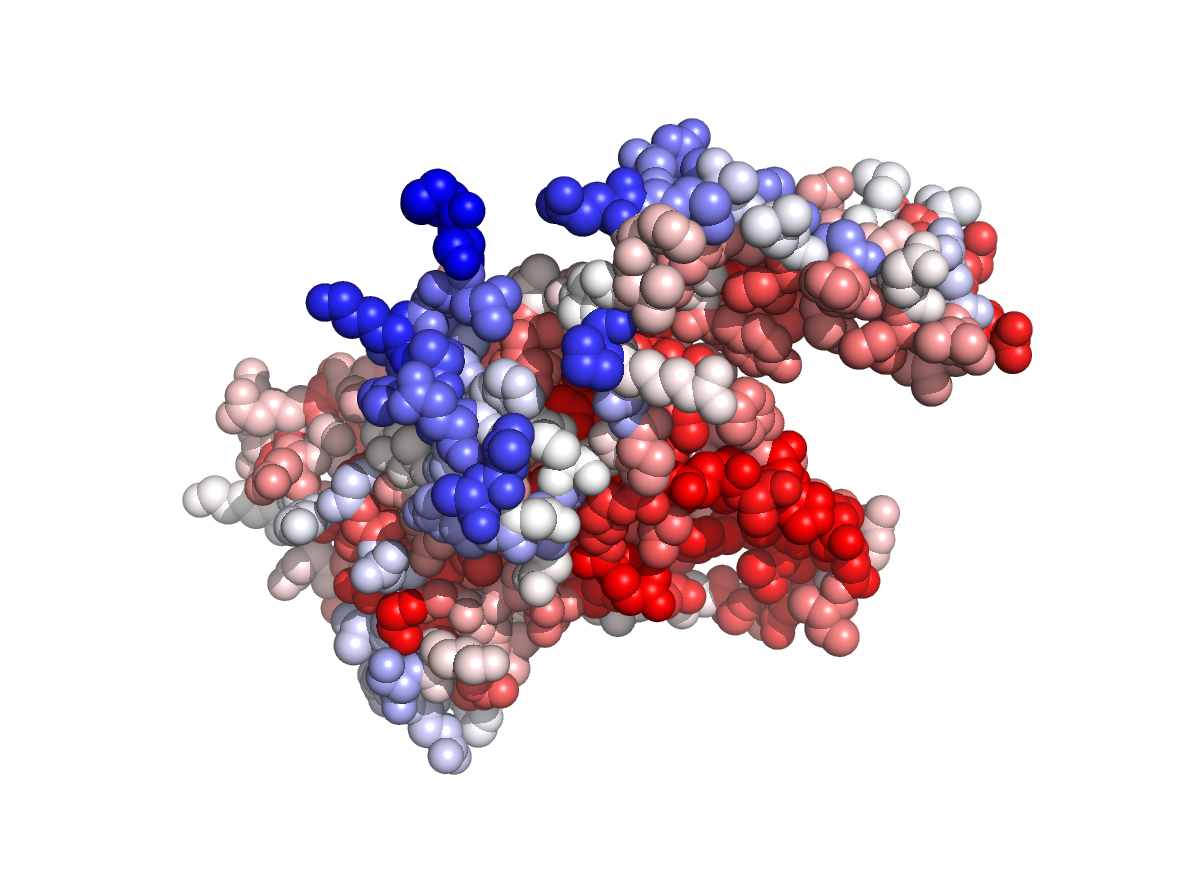 |
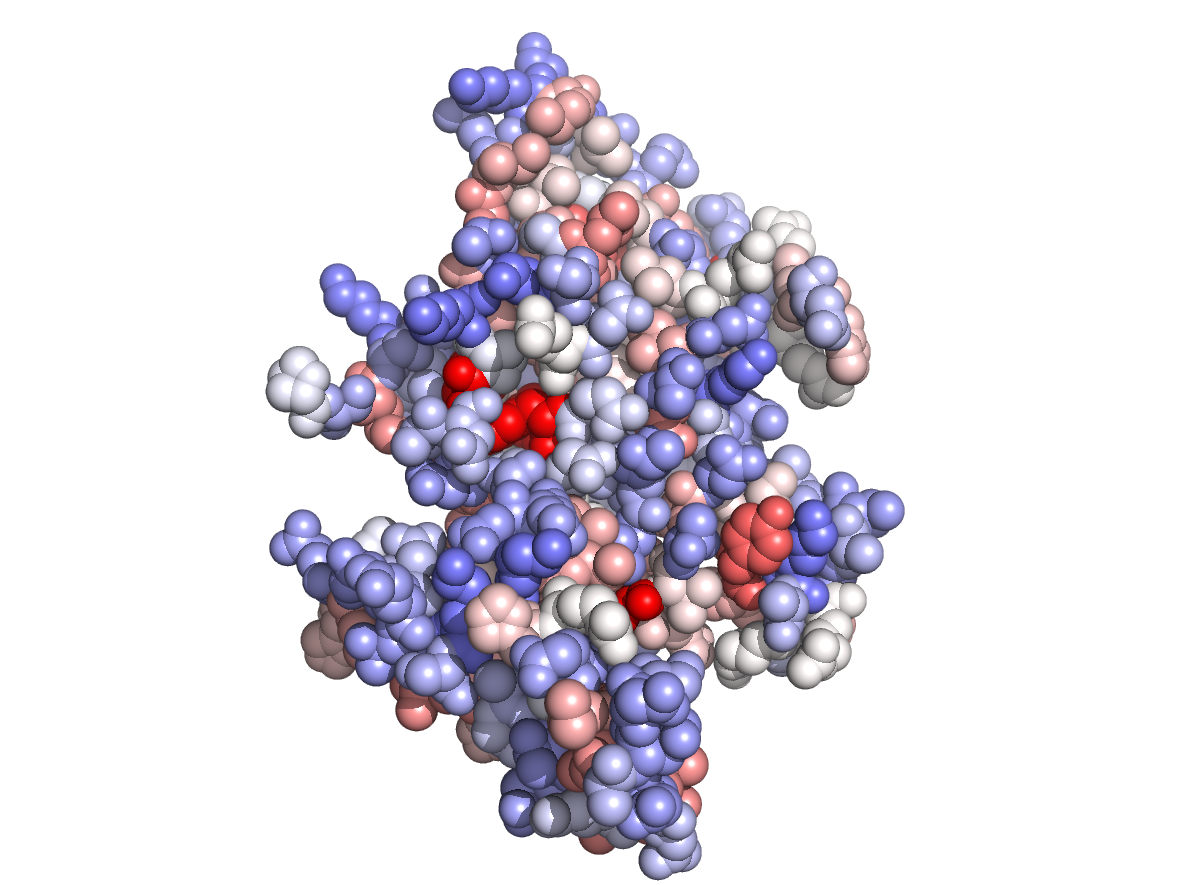 |
 |
PDB Structure: 1A8R chain O
Predicted sites - SC1 |
Predicted sites - SC2 |
Predicted sites - SC3 |
 site |
 site |
 site |
Evolutionary conservation - TJET |
Physico-chemical properties - PC |
Circular variance - CV |
 |
 |
 |
